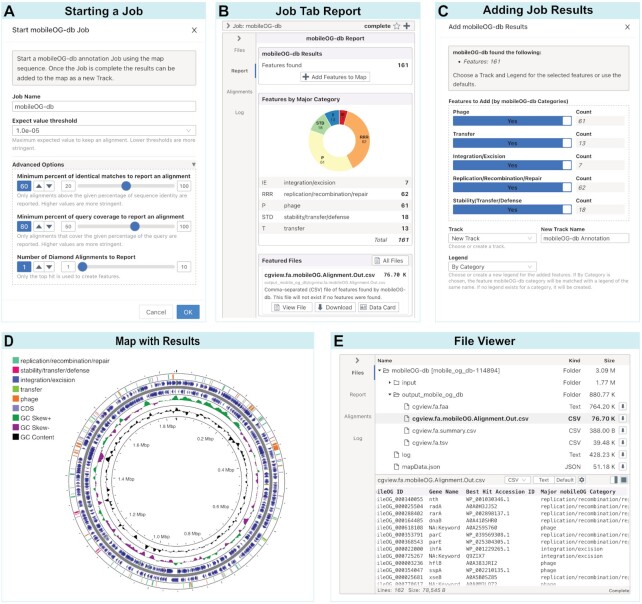
Figure 4.
Server-based tool workflow. The mobileOG-db tool is shown as an example. (A) Starting a server-based tool will show the Start Dialog where the name for the job can be provided as well as any tool-specific options. (B) Completed jobs display a report card with a summary of features found and a button to add them to the map. The report also includes a list of featured files (i.e. key results files) with links to view or download each file. (C) Add Dialog for adding job results to the map with options for selecting which features to add and which track and legend to use for the added features. (D) Map with added features. Shown are the original features (i.e. CDS, tRNA, rRNA) extracted from a GenBank file (NZ_CP007470), the mobileOG-db features split into five categories (e.g. stability/transfer/defense, replication/recombination/repair, integration/excision, transfer and phage) and the results of the GC Content and GC Skew tool. (E) File Card showing the file tree of input and output files for this job (top) and the file viewer for one of the output files (bottom).