Abstract
Objectives
Clinical notes are a veritable treasure trove of information on a patient’s disease progression, medical history, and treatment plans, yet are locked in secured databases accessible for research only after extensive ethics review. Removing personally identifying and protected health information (PII/PHI) from the records can reduce the need for additional Institutional Review Boards (IRB) reviews. In this project, our goals were to: (1) develop a robust and scalable clinical text de-identification pipeline that is compliant with the Health Insurance Portability and Accountability Act (HIPAA) Privacy Rule for de-identification standards and (2) share routinely updated de-identified clinical notes with researchers.
Materials and Methods
Building on our open-source de-identification software called Philter, we added features to: (1) make the algorithm and the de-identified data HIPAA compliant, which also implies type 2 error-free redaction, as certified via external audit; (2) reduce over-redaction errors; and (3) normalize and shift date PHI. We also established a streamlined de-identification pipeline using MongoDB to automatically extract clinical notes and provide truly de-identified notes to researchers with periodic monthly refreshes at our institution.
Results
To the best of our knowledge, the Philter V1.0 pipeline is currently the first and only certified, de-identified redaction pipeline that makes clinical notes available to researchers for nonhuman subjects’ research, without further IRB approval needed. To date, we have made over 130 million certified de-identified clinical notes available to over 600 UCSF researchers. These notes were collected over the past 40 years, and represent data from 2757016 UCSF patients.
Keywords: clinical note text, de-identification, unstructured data, Philter
BACKGROUND AND SIGNIFICANCE
The field of precision medicine is quickly generating new approaches to disease treatment that are customized for patients based on their personal genetics, medical history, lifestyle, and social determinants of health. For targeted therapies to advance, there is a growing need for in-depth patient data beyond structured electronic health records (EHR). Free-text clinical notes contain detailed accounts of a patient’s medical and family history, lifestyle, disease progression, treatment plans, doctor sentiments and prognoses, which are not typically captured in structured EHR data but are highly valuable for personalized disease treatment research.1,2 For example, de-identified clinical notes proved extremely valuable during the COVID-19 pandemic, when many investigators wanted to quickly advance COVID-19 research without having to undergo several months of Institutional Review Board (IRB) reviews.3,4 Unfortunately, clinical notes remain largely unexplored in precision medicine research studies due to the presence of protected health information (PHI) and access restrictions posed by IRBs.
In large medical centers like the University of California, San Francisco (UCSF), about 2 million new clinical notes are generated monthly, thus steadily building the current corpus of 130 million notes (as of January 2023). The need for a fast and robust pipeline that can reliably de-identify millions of clinical notes at once is invaluable. However, off-the-shelf tools and software to redact PHI fall short on de-identification quality and require significant effort to scale for large volumes of data. A previous open-source version of Philter5 using file Input/Output (I/O), presented various scaling challenges. Using that version, notes extracted from the UCSF’s instance of Epic Clarity, a Third Normal Form (3NF) data warehouse and relational database for clinical data, had to be reconstructed into individual text files and stored in subdirectories to accommodate file count limits in Linux’s XFS filesystem. Computing the monthly change in data is also suboptimal when using millions of files. Parallel processing across multiple servers posed network overload issues with file I/O. For example, a batch of 1000 clinical notes took up to an hour to process. Existing de-identification tools and algorithms like Physionet,6 Scrubber,7 and open-source Philter do not or insufficiently address the following needs:
Scalability for large-scale, iterative de-identification of hundreds of millions of clinical notes.
Qualitative and quantitative certification of the algorithm and the de-identified notes, as per Health Insurance Portability and Accountability Act (HIPAA) Privacy Rule de-identification standards, for usage without further IRB restrictions.
Enhanced features like date obfuscation, where date fields are shifted to mimic the timing of clinical events without revealing actual PHI.
OBJECTIVES
The main goals are to create an automated version of the Philter (V1.0) de-identification process for unstructured clinical notes and to make them available as a certified de-identified asset of UCSF’s Information Commons data-science platform.8 Philter V1.0 improves upon the original open-source de-identification software Philter Beta,5 specifically, by addressing the following needs:
-
Algorithmic features:
Capture all patient names that are also common English words.
Capture concatenated PHI and partial addresses.
Rescue valid pathology and genomic terms, which were redacted in the original version.
Redact small-town names with populations of less than 30000 to reduce the possibility of reidentification.
Provide additional features like date shifting.
-
Processing features:
Accelerate the de-identification process by using a MongoDB, instead of traditional file I/O. The need for rapid access to clinical notes during the COVID-19 pandemic highlighted the need for faster note processing.
Establish a certified pipeline in accordance with HIPAA’s Privacy Rule via external audit.
With these enhancements, the Philter V1.0 pipeline is a completely certified, scalable solution for de-identifying unstructured textual data. This pipeline has been tested, verified, and implemented at UCSF, and provides researchers with monthly clinical notes refreshes that are made available through secure search engines like EMERSE9 and Apache Solr. These notes are made available with no IRB restrictions to UCSF investigators, thus supporting rapid access to rich data sources that can be used to advance clinical research.
MATERIALS AND METHODS
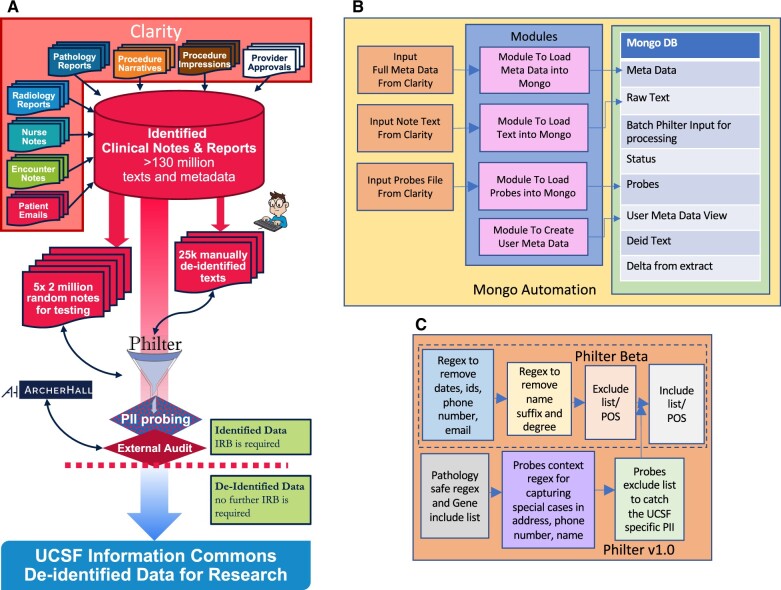
One of the main goals of developing Philter V1.0 was to establish a certified de-identification pipeline in accordance with the HIPAA Privacy Rule (Figure 1A). There are 2 methods that can be used to satisfy the Privacy Rule’s de-identification standards: Expert Determination and Safe Harbor. We engaged the expertise of ArcherHall, a data forensics company, to determine which de-identification method should be used to certify different parts of the pipeline. During the certification process, we made several feature enhancements to the Philter Beta code that improved de-identification to a point where both the UCSF security panel and ArcherHall were able to approve the algorithm, the associated pipeline, and the de-identified clinical notes as certified de-identified using both programmatic and manual verification processes. In addition, we replaced the file storage with MongoDB and used multithreading to accelerate the de-identification process on a large and diverse corpus of clinical notes with over 150 note types.
Figure 1.
Schematic of Philter V1.0 pipeline. (A) Complete pipeline, (B) MongoDB automation, and (C) Philter V1.0 algorithm features.
Certification process
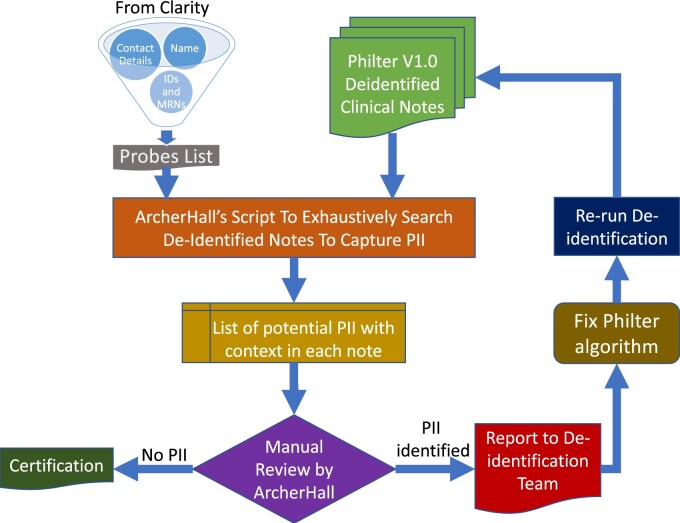
Clinical notes redacted using Philter V1.0 underwent comprehensive security testing by ArcherHall, who reviewed the algorithm and performed data validation to determine whether any personal identifying information (PII) remaining in de-identified notes presented little to no risk of reidentification. During each round of this iterative process, a batch of random 2M de-identified clinical notes was assessed by ArcherHall through their in-house testing algorithm (Figure 2). Their algorithm searched our de-identified clinical notes for more than 70 million PIIs extracted from the UCSF Epic Clarity and Caboodle databases. Their search was performed on each of the 5 independent test sets of 2M notes we provided after each algorithmic enhancement detailed in the “Materials and Methods” section, respectively. Each iteration resulted in a list of all potential PIIs that persisted in each of the 2M notes test sets. ArcherHall then performed extensive manual checks and reviews on this result to compile a list of true positive lingering PII. The list was then used to improve the Philter algorithm accordingly. After multiple such iterations of the process described above, a total of 10M de-identified clinical notes was provided to ArcherHall for certification.
Figure 2.
ArcherHall certification process.
To ensure the de-identified clinical notes were HIPAA compliant the “Safe Harbor” criterium was applied to all PHI types except for dates to verify that no lingering PII remained, and the “Expert Determination” method for de-identified date fields. For date fields, ArcherHall verified that appropriate processing was performed to obfuscate the original date and determined the risk of reidentification using publicly available information, including social media posts. ArcherHall determined through the programmatic and manual verification process that there are no lingering PII in the de-identified data and the potential for reidentification of patients is extremely low. Thus, ArcherHall issued a certificate stating the de-identified clinical notes and the pipeline used to produce the de-identified data meet the HIPAA de-identification criteria.
Philter V1.0 algorithm enhancements toward certification
The Philter algorithm uses a set of rules and statistical methods to distinguish between PHI and “safe” (non-PHI) words. Despite Philter Beta having the highest overall recall when compared to its 2 strongest competitors at the time, Physionet and Scrubber, there were known issues with precision because Philter flags any token that is not explicitly marked as PHI or non-PHI as putative PHI.6,10 When developing Philter V1.0, we implemented select enhancements to improve overall performance without compromising on either precision or recall. Each of the algorithmic enhancements was done based on the results produced by ArcherHall after extensive programmatic and manual review of our de-identified clinical test sets. Philter uses a set of regular expression exclusion patterns (ie, patterns and lists of words known to be PHI in most instances) to flag PHI tokens, and “safe” regular expressions (ie, patterns that detect a token or word known not to be PHI) and manually curated include lists (ie, lists of words that are known not to be PHI in the vast majority of instances, eg, common English words) to flag non-PHI tokens. We implemented the following algorithmic changes to improve upon the previous Philter version (Figure 1C). Each algorithmic change was given a new version tag to distinguish them from the previous version and was tested on a new set of random 2M clinical notes since many features were implemented to capture rare cases of PHI.
Date Shifting (Philter Delta): In the previous Philter version, all dates were obfuscated. While this approach was effective for improving recall, it did not allow researchers to analyze clinically relevant timelines. To retain relative timelines while obscuring PHI, Philter V1.0 shifts all dates according to a fixed offset (−365 day to −1 day) that is randomly generated for each patient, and then normalizes each detected date into a Python Datetime object. If the normalized date does not contain a fully defined year, month, or day, the missing date components are inferred using previously defined defaults. After the full Datetime object is shifted, only the date components that were present in the original text are added to the de-identified text.
Capturing names that are common English words (Philter Epsilon): In the previous Philter version, common English words were removed from name exclude lists generated from 2010 US census data to improve precision. However, this allowed some PHI to escape obfuscation. To address this problem, a dynamic exclude list was implemented to capture first or last names that are also common English words (eg, “Long,” “Field,” “May”). This feature leverages a “Probes” file, which is built using personally identifiable information (PII) data like names, phone number, addresses, social security numbers from Clarity for all 2.75M UCSF patients. Probes are tokenized on whitespace and special characters, converted to lowercase, and passed as an optional input to the algorithm. This feature also leverages the Python NLTK Parts of Speech (POS) tagging module11 to capture only NNPs (proper nouns). The POS module uses statistical methods to determine the structure of each sentence to reduce false positives when capturing names known via the Probes. When Philter V1.0 processes a clinical note, it dynamically generates a unique list of labeled PII associated with that patient from the Probes file (eg, first name(s), last name(s), addresses, medical record numbers, etc.), which is added to Philter’s set of obfuscation rules. A word is obscured in the text file if it is tagged as NNP by the POS tagger and appears in the list of names for that patient. If no PII exists in Clarity for a particular patient, this step is skipped. The success of the name dynamic exclude list motivated us to expand this feature to other types of PII like telephone numbers, zip codes, addresses, and workplace.
90+ age obfuscation (Philter Eta): The HIPAA Privacy Rule requires that all data from patients older than 90 years be obfuscated. For these patients, Philter V1.0 shifts all birth dates such that they appear to be at most 90 years old on the day of data extraction, or on the date of death for deceased patients. Deceased patients are identified using both UCSF records and the California Death Registry. Ages >90 years are obfuscated (not shifted) to maintain consistency.
Rescuing Pathology and Gene Terminologies (Philter Theta): Philter Beta over-redacted some clinically important terms like gene names and pathology terms as the parts-of-speech tagging falsely identified these terms as proper nouns which caused over redaction. At the request of several researchers from UCSF, we enhanced the algorithm to rescue some of these terms. We compiled a list of “safe” pathology terms (like lymph node numbers and cassette numbers) from 3 million pathology reports and identified more than 43 000 common gene symbols from NCBI such as BRCA1, A1BG. We then incorporated these lists into the algorithm as a new include list so that these terms could be retained in the de-identified notes.
Capturing Alphanumeric Names, Partial Addresses, Phone Numbers, and Patient Workplace (Philter Iota): In some cases, the token-level dynamic exclude list did not capture patient names that were concatenated with numeric sequences (eg, “1-1-1bill”). To capture these edge cases, we developed a “dynamic regular expression” feature that combined name probes and digit matchers into the same regular expression. For example, the patient name “bill” would be compiled into a regular expression that searches for a variety of numeric patterns directly concatenated with the string “bill”, such as “1.1.1bill”, “1/2/3bill”, “1000-2000-3000bill”, etc. We employed a similar technique to capture partial instances of phone numbers, addresses, and patient workplace, but instead combined these probes with searches for special characters and blank spaces. For example, the area code “111” would be compiled into the regular expression r’111[^0-9A-Za-z]{1,5})?, which searches for the string “111” followed by 1–5 special characters. We also implemented a “dynamic context regular expression” feature that helped make use of single letters from the name probes (initials or part of a first name, eg, “M.K.”) without sacrificing precision. This feature checks for name PHI (identified upstream in the pipeline) that flanks a probe-matching token on one or both sides. If the token is indeed flanked by name PHI, the token is obscured. For example, consider the phrase “Jane. A. Doe.” If the tokens “Jane” and/or “Doe” were marked as PHI upstream in the pipeline, the dynamic context regular expression feature would obscure the letter “A” if it were also in the list of name probes for that note. Note that these are rare edge cases which are caused primarily due to typos.
Small town names (Philter V1.0): Small-town names (population <20000) in the clinical notes poses a risk for reidentification of patients from those towns. To address this issue, we created an exclude list of town names with population <30000 from the US census registry (as of 2019).
Improved scalability and performance
Clinical notes are prepared for de-identification using an automated pipeline, which loads notes and metadata into MongoDB and performs quality assurance checks against the previous notes extract from Clarity. The MongoDB collections for each notes extract are shown in Figure 1B.
The automated pipeline has 8 main modules that use our Notes extract, transform, load process during each data refresh:
Extract clinical notes texts and corresponding metadata from Clarity.
Load metadata, date offsets (for date shifting), and ID mappings (for original to surrogate ID crosswalks) extracted from the Clinical Data Warehouse into the Meta Data collection.
Load texts corresponding to the metadata into the Raw Text collection.
Load PII data extracted from Clarity into the Probes collection.
Create a Batch list of notes for de-identification for the current refresh. This includes the subset of notes added to Clarity and the notes that were modified since the previous refresh.
Run Philter V1.0 on notes in the Batch list and save the de-identified notes in the Deid Text collection.
Create the Meta Data View collection with only de-identified data for users by hiding original identifiers and date offsets from the metadata.
Transfer the de-identified clinical note texts and user-facing metadata to Information Commons.
UCSF maintains a HIPAA-compliant PHI-safe compute environment that is implemented on the VMware Virtual Platform (vSphere). The system consists of one VMware virtual Linux CentOS server which hosts a MongoDB database configured with 48 vCPU, 412GB RAM, and 5TB vSAN storage drive, and 4 worker servers for Philter batch processing with 32 vCPU and 256GB of RAM each. All communication between the 5 servers is encrypted during the Philter V1.0 de-identification process. Philter V1.0 can launch the de-identification process on all 5 servers using Secure Shell protocol. Once the notes are processed, the de-identified clinical note text along with the user-facing metadata are securely transferred to our Information Commons platform12 and to a Microsoft SQL server hosting the certified de-identified data for researchers.
RESULTS
Philter V1.0 streamlines and automates the clinical notes de-identification process and makes it scalable for large corpora of unstructured text data. The algorithmic enhancements made to the previous version of Philter and the certification techniques employed by ArcherHall improved the performance of Philter and led to professionally certify a large notes corpus as de-identified.
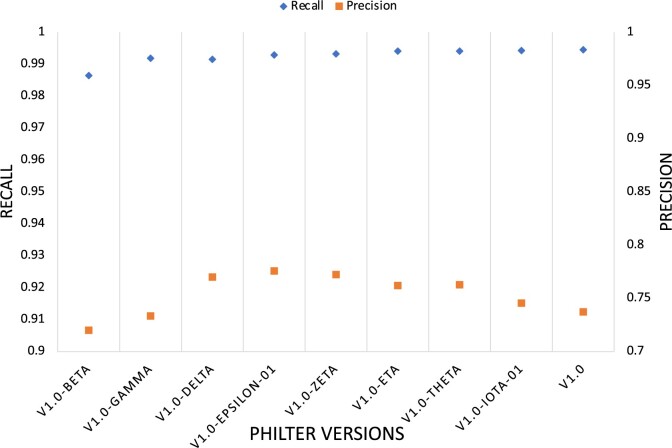
To ensure that the high precision and recall we had already achieved was preserved, we saved each enhancement as a version and tested on 18000 test notes from the gold standard corpus. We observed minimal impact on the precision and recall on the manually annotated 18000 test set (Figure 3).
Figure 3.
Precision and recall over each Philter iteration on 18 000 manually annotated notes.
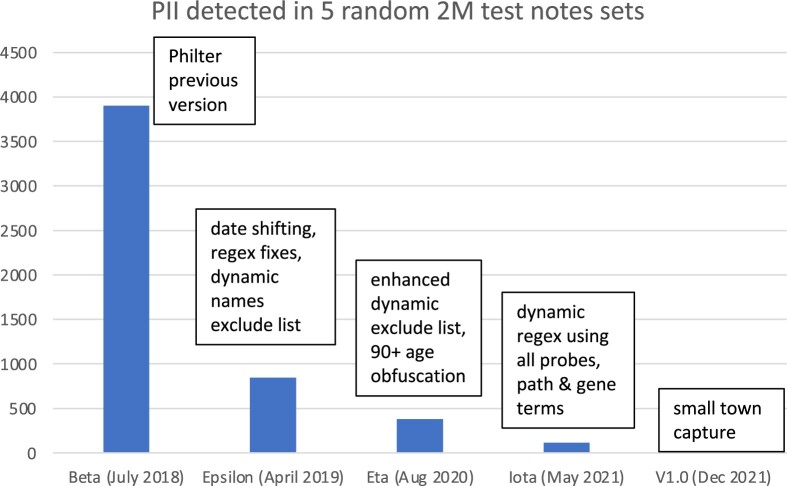
Our original manually annotated corpus of 25000 notes set aside for testing and training purposes was not sufficient for evaluating the true impacts of the Philter V1.0 enhancements since we were intent on capturing rare edge cases of PHI. We therefore used 100000 random clinical notes to train the algorithm and tested on batches of 2 million random notes which were provided to ArcherHall for further testing. With each new batch of 2M test notes, we iteratively improved our algorithm reducing PII until none persisted (Figure 4). A total of 10M clinical notes (ie, 5 batches of 2M notes) were programmatically and manually reviewed by ArcherHall to determine that there were no instances of PII.
Figure 4.
Personal identifying information (PII) detected by ArcherHall after each development cycle in a newly drawn random test set of 2 million notes.
The main enhancements yielded the following results:
Philter Delta: Normalized shifted dates were made available in clinical notes to meet research use cases that require consistent relative event dates.
Philter Epsilon: In the 100000 notes set, the dynamic exclude list captured 1294 additional names tokens that were common names like Long, Black, Short, etc., and incorrectly marked only 256 tokens like no, none, baby, study, health, etc. as PHI. The false positives were primarily due to bad probes in the data extracted from Clarity. We also identified that implementing this feature together with improving some existing regular expressions reduced the number of remaining PII from 3900 in a random 2M notes test set to 848 tokens in the next iteration.
Philter Eta: By shifting all birth dates for patients older than 90 years such that they appear to be 90 years old, we ensured that the data produced by our de-identification pipeline is HIPAA compliant. This special handling of geriatric patients’ records and further refinement of the dynamic exclude list resulted in a reduced number of 380 PII in the next 2M random notes set.
Philter Theta: After implementing the gene and pathology include list, we were able to rescue ∼4000 gene names like APOA1, CYP27A1, and pathology terms like cassette numbers and molecular markers like MLH1, PMS2 in our training set of 100000 notes without compromising recall or precision in our test sets (Figure 3). Based on this observation we estimate that more than 5 million gene names have been rescued in our 130 million clinical notes, thus improving the usefulness for research.
Philter Iota: Capturing alphanumeric names like “123Sam”, partial addresses, phone numbers, workplace, using dynamic context regular expressions we found that many additional PII tokens were captured lowering the remaining PII to 112 tokens in an independent random 2M test set.
Philter V1.0: Small-town names like Lucerne (pop 2896), Bay Point (pop 25808), Larkspur (pop 12319) were removed from the clinical notes as ArcherHall expressed concern due to their low population. By implementing this final fix of the algorithm we were able to remove all lingering PII from the 10 million clinical notes corpus as validated by ArcherHall.
ArcherHall determined that for the date obfuscation, the risk of reidentification using publicly available data, including social media posts, was extremely low, with less than 0.025% of patients at risk for reidentification. They found no instances of the 17 nondate fields (names, geographic location, telephone number, vehicle identifier, fax number, email, etc.) remained in the clinical note text, as required by the “Safe Harbor” method. Analysis of the user-facing metadata returned only false positive results, which were manually confirmed by ArcherHall. Philter V1.0 is a rule-based algorithm with a series of regular expressions and include/exclude lists implemented in a particular order on the clinical notes to redact them. The certification is specific to UCSF and the version of Philter used to de-identify this data. Any changes to the algorithm or introduction of new textual data sources must undergo a re-certification similar to the certification process described above. The certificate issued by ArcherHall needs to be renewed every 2 years to ensure that the de-identified clinical notes and the pipeline continue to be HIPAA compliant.
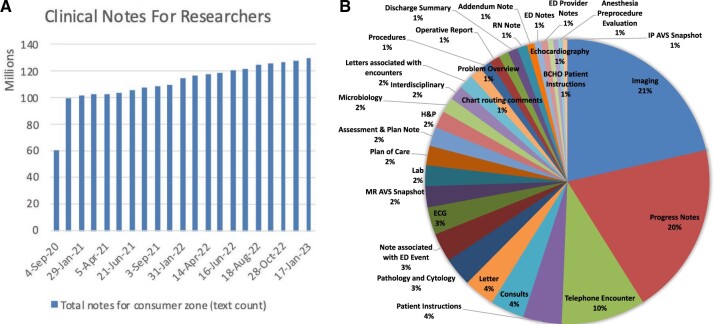
MongoDB data storage and multiserver parallel processing improved Philter scalability immensely. By using MongoDB, instead of traditional file I/O, we observed a 6-fold improvement in scalability. The pipeline can now process an average of 13M notes (in batches of 1000, spread across 4 computational nodes, with 30 parallel threads per node) under 24h. This brings our processing time for 130 million clinical notes down from several months to mere weeks. The full clinical notes de-identification pipeline is now operational at UCSF and is on a regular refresh cycle with about 2M new clinical notes per month added to the Information Commons (Figure 5A). These notes span over 150 different note types (Figure 5B) and have been vital in supporting cutting edge research studies in gastroenterology,13 radiology,14 and pulmonology15—to name a few.
Figure 5.
(A) Monthly refresh of clinical notes for research and (B) distribution of available types of de-identified clinical notes.
SIGNIFICANCE AND USE CASES
Several research projects at UCSF have benefitted from the availability of periodically updated, de-identified clinical note texts. At UCSF, we have set up Apache cTAKES16 in a high-throughput client-server model (cTAKES-HT),17 which is a fast natural language processing pipeline for extracting clinically relevant information from de-identified clinical text. All 130 million clinical notes have been processed through cTAKES-HT, extracting more than 5 billion medical concepts. The de-identified notes and extracted concepts have been made available to more than 600 researchers at UCSF. Some of the research projects that have used this data resource and successfully published their findings include:
Accurate Machine Classification of Ulcerative Colitis: Mayo Subscores from Electronic Health Record Procedure Reports13
Identifying Patients with Interstitial Lung Disease in Electronic Health Records: Development and Validation of Machine Learning Algorithms15
Federated learning for predicting clinical outcomes in patients with COVID-1914
Other projects in progress include a hip fracture detection study, an evaluation of differential diagnoses and patient similarity in neurodegenerative conditions, and other studies in the areas of social determinants of health, pathology, and oncology.
CONCLUSION
We have established an automated clinical text de-identification pipeline at our institution that is HIPAA compliant, and scalable to millions of clinical texts. The entire audit and development of Philter V1.0 took several years. The work started in 2016, with the release of the open-source version in 2018, and iterated refinement to the final certificate issued in 2021.To the best of our knowledge, Philter V1.0 is currently the only professionally certified de-identification software for unstructured data. This algorithm is now being adopted by other medical centers, including the University of California, Irvine and the University of California, Davis.
The certified de-identified clinical note texts are available to the UCSF research community without the need for any further IRB approval. There are, however, access requirements in place, consistent with an existing approved process for accessing de-identified data assets. These include: (1) Statement of Responsibility for Requestor of Personally Identifiable Information and/or Protected Health Information; (2) Acknowledgement of Liability for the Use of UCSF Enterprise De-identified Data Sources; (3) HIPAA and UCSF cybersecurity training; and (4) PI approvals for team members.
Although Philter V1.0 is already a certified de-identification tool, we continue to actively develop and improve the algorithm to address redaction issues or feature enhancement requests by our data users. Efforts are in progress to improve redaction errors related to numeric ranges being misinterpreted as dates, and other over-redaction errors of potentially informative medical terms and phrases. The de-identification pipeline and the data need to be recertified every 2 years to ensure that the data and the algorithm continue to be HIPAA compliant. Any new type of clinical text beyond the certified 150 note types needs to be certified following all the same certification procedures before they can be added as part of the certified de-identified data asset.
ACKNOWLEDGMENTS
The work was done under IRB No. 16-20784. The certificate and audit were done by ArcherHall. We would like to express our gratitude for the collaborations with UCSF IT Academic Research Systems, UCSF Privacy Office, and UCSF IT Governance Enterprise Information and Analytics Committee. We are also grateful to all of the many staff and student trainees who contributed programming code to this project via the Bakar Computational Health Sciences Institute and OptumLabs Data Science Internship Program at UCSF.
Contributor Information
Lakshmi Radhakrishnan, Academic Research Services, Information Technology, University of California, San Francisco, San Francisco, California, USA.
Gundolf Schenk, Bakar Computational Health Sciences Institute, University of California, San Francisco, San Francisco, California, USA.
Kathleen Muenzen, Bakar Computational Health Sciences Institute, University of California, San Francisco, San Francisco, California, USA.
Boris Oskotsky, Bakar Computational Health Sciences Institute, University of California, San Francisco, San Francisco, California, USA.
Habibeh Ashouri Choshali, Bakar Computational Health Sciences Institute, University of California, San Francisco, San Francisco, California, USA.
Thomas Plunkett, ArcherHall LLC, Sacramento, California, USA.
Sharat Israni, Bakar Computational Health Sciences Institute, University of California, San Francisco, San Francisco, California, USA.
Atul J Butte, Bakar Computational Health Sciences Institute, University of California, San Francisco, San Francisco, California, USA; Department of Pediatrics, University of California, San Francisco, San Francisco, California, USA; Center for Data-Driven Insights and Innovation, University of California Health, Oakland, California, USA.
FUNDING
The work has been funded intramurally at UCSF, 2016 to date, with a combination of funds from Bakar Computational Health Sciences Institute and the Marcus Foundation Grant for Precision Medicine.
AUTHOR CONTRIBUTIONS
LR has written the manuscript and is the main developer of the algorithmic improvements and the pipeline. GS has revised the manuscript and led the development of the algorithm, the pipeline, and the approval process. KM is a codeveloper of many of the algorithmic enhancements. BO is the system administrator. HAC tested the algorithm and made plots. TP performed the expert reviews for the certification of the de-identification process and resulting data. SI and AJB have advised and managed the work. All authors have critically reviewed the manuscript, approved it for publication, and agree to be accountable for all aspects of the work.
CONFLICT OF INTEREST STATEMENT
AJB is a cofounder and consultant to Personalis and NuMedii; consultant to Mango Tree Corporation, and in the recent past, Samsung, 10x Genomics, Helix, Pathway Genomics, and Verinata (Illumina); has served on paid advisory panels or boards for Geisinger Health, Regenstrief Institute, Gerson Lehman Group, AlphaSights, Covance, Novartis, Genentech, and Merck, and Roche; is a shareholder in Personalis and NuMedii; is a minor shareholder in Apple, Meta (Facebook), Alphabet (Google), Microsoft, Amazon, Snap, 10x Genomics, Illumina, Regeneron, Sanofi, Pfizer, Royalty Pharma, Moderna, Sutro, Doximity, BioNtech, Invitae, Pacific Biosciences, Editas Medicine, Nuna Health, Assay Depot, and Vet24seven, and several other nonhealth related companies and mutual funds; and has received honoraria and travel reimbursement for invited talks from Johnson and Johnson, Roche, Genentech, Pfizer, Merck, Lilly, Takeda, Varian, Mars, Siemens, Optum, Abbott, Celgene, AstraZeneca, AbbVie, Westat, and many academic institutions, medical or disease specific foundations and associations, and health systems. AJB receives royalty payments through Stanford University, for several patents and other disclosures licensed to NuMedii and Personalis. AJB’s research has been funded by NIH, Peraton (as the prime on an NIH contract), Genentech, Johnson and Johnson, FDA, Robert Wood Johnson Foundation, Leon Lowenstein Foundation, Intervalien Foundation, Priscilla Chan and Mark Zuckerberg, the Barbara and Gerson Bakar Foundation, and in the recent past, the March of Dimes, Juvenile Diabetes Research Foundation, California Governor’s Office of Planning and Research, California Institute for Regenerative Medicine, L’Oreal, and Progenity. The authors have declared that no competing interests exist.
DATA AVAILABILITY
The data underlying this article were accessed from UCSF electronic medical records and cannot be shared publicly due to the patients’ privacy. The derived data and software generated from this research will be shared on reasonable request to the corresponding author with permission of the UCSF IT Governance.
REFERENCES
- 1. Cefalu WT, Andersen DK, Arreaza-Rubín G, et al. Heterogeneity of diabetes: β-cells, phenotypes, and precision medicine: Proceedings of an International Symposium of the Canadian Institutes of Health Research’s Institute of Nutrition, Metabolism and Diabetes and the U.S. National Institutes of Health’s. Diabetes Care 2022; 45 (1): 3–22. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Sirota M, Thomas CG, Liu R,. et al. Enabling precision medicine in neonatology, an integrated repository for preterm birth research. Sci Data2018; 5: 180219. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Dyrbye LN, Thomas MR, Mechaber AJ, et al. Medical education research and IRB review: an analysis and comparison of the IRB review process at six institutions. Acad Med 2007; 82 (7): 654–60. [DOI] [PubMed] [Google Scholar]
- 4. Liberale AP, Kovach JV.. Reducing the time for IRB reviews: a case study. J Res Admin 2017; 48: 37–50. [Google Scholar]
- 5. Norgeot B, Muenzen K, Peterson TA,. et al. Protected Health Information filter (Philter): accurately and securely de-identifying free-text clinical notes. NPJ Digit Med2020; 3: 57. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Goldberger AL, Amaral LA, Glass L,. et al. PhysioBank, PhysioToolkit, and PhysioNet: components of a new research resource for complex physiologic signals. Circulation2000; 101 (23): E215–20. [DOI] [PubMed] [Google Scholar]
- 7. McMurry AJ, Fitch B, Savova G, Kohane IS, Reis BY.. Improved de-identification of physician notes through integrative modeling of both public and private medical text. BMC Med Inform Decis Mak2013; 13: 112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. UCSF. UCSF DeID CDW. Data set. San Francisco: University of California, Academic Research Systems; 2022. Report No.: R20220207.
- 9. Hanauer DA. EMERSE: the electronic medical record search engine. AMIA Annu Symp Proc 2006; 2006: 941. [PMC free article] [PubMed] [Google Scholar]
- 10. Aberdeen J, Bayer S, Yeniterzi R,. et al. The MITRE Identification Scrubber Toolkit: design, training, and assessment. Int J Med Inform2010; 79 (12): 849–59. [DOI] [PubMed] [Google Scholar]
- 11. Toutanova K, Klein D, Manning CD, Singer Y. Feature-rich part-of-speech tagging with a cyclic dependency network. In: Proceedings of the 2003 Human Language Technology Conference of the North American Chapter of the Association for Computational Linguistics; 2003: 252–9. [Google Scholar]
- 12. Schenk G. UCSF Information Commons: multifactor data science to advance precision medicine. 2022. https://www.pmwcintl.com/previous/2022sv/. Accessed June 2022.
- 13. Rudrapatna V, Gupta S, Mardirossian T, Narain R, Mosenia A, Butte A.. Accurate machine classification of ulcerative colitis Mayo subscores from electronic health record procedure reports. Am J Gastroenterol 2020; 115 (1): S420. [Google Scholar]
- 14. Dayan I, Roth HR, Zhong A, et al. Federated learning for predicting clinical outcomes in patients with COVID-19. Nat Med 2021; 27 (10): 1735–43. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Farrand E, Gologortskaya O, Radhakrishnan L, Mills H, Collard HR, Butte A.. Identifying Patients with Interstitial Lung Disease in Electronic Health Records: Development and Validation of Machine Learning Algorithms [Conference Presentation]. Chicago, IL: AMIA; 2022. [Google Scholar]
- 16. Savova GK, Masanz JJ, Ogren PV, et al. Mayo clinical text analysis and knowledge extraction system (cTAKES): architecture, component evaluation and applications. J Am Med Inform Assoc 2010; 17 (5): 507–13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Abramowitsch P. Apache cTAKES High Throughput Orchestration [Conference Presentation]. ApacheCon; 2020.https://www.apachecon.com/acah2020/tracks/ctakes.html. [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Data Availability Statement
The data underlying this article were accessed from UCSF electronic medical records and cannot be shared publicly due to the patients’ privacy. The derived data and software generated from this research will be shared on reasonable request to the corresponding author with permission of the UCSF IT Governance.