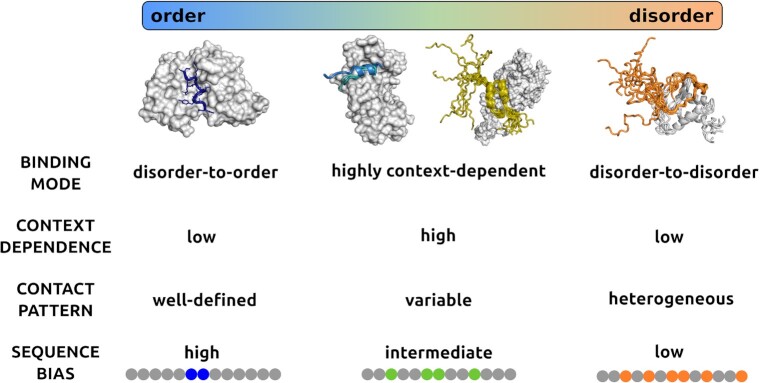
Figure 1.
Binding modes of protein interactions. Protein interactions sample a wide range of binding modes from ordered to disordered binding. The different binding modes are characterised by structural order or disorder in the bound state (horizontal bar), which are reflected by the heterogeneity of contact patterns. In ordered binding the contact patterns are well-defined, while in disordered binding they are heterogeneous, and in case of context-dependent binding, they can be either well-defined or variable depending on the cellular conditions. At the ends of the binding mode spectrum, proteins preferentially sample a unique binding mode, therefore the context dependence is weak. In the central region of the spectrum, protein regions can sample both ordered and disordered binding modes, so the context-dependence is high. Ordered binding is generated by a strong local sequence bias (blue), whereas disordered binding is due a weak sequence bias (orange). In context-dependent binding modes, the sequence bias is modulated by the conditions, and can be strong or weak depending on the context (green). Ordered binding modes, represented by the complex of merozoite surface protein 2 (blue, top left) from Plasmodium falciparum and the monoclonal antibody m6D8 (gray, PDB:4qxt (41)). Disordered binding is represented by the interactions between leukemia fusion protein AF9 (gray) and elongation factor AF4 (orange) (PDB:2lm0 (42), top right), where both partners retain considerable conformational heterogeneity in the bound complex. Context-dependent binding can be achieved by polymorphism, i.e. adopting different secondary structures upon binding such as in the case of ribosomal S6 kinase 1 (marine, slate, second to left) binding to S100b (gray, PDB:5csf, 5csi, 5csj (43)) or retaining conformational disorder in a condition-dependent manner such as in the case of p150 subunit of the eukaryotic initiation factor 4F (yellow) binding to the translation initiation factor 4E, (gray PDB: 1rf8 (44)).