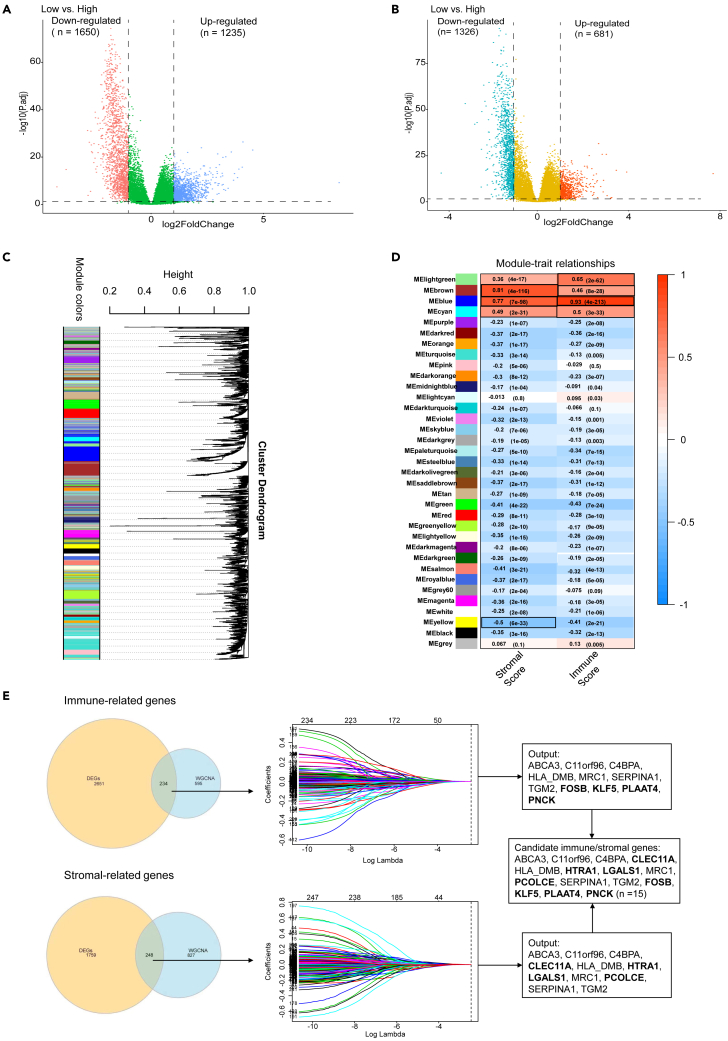
Figure 2.
Identification of significant TME-related genes
(A and B) The volcano map shows the differentially expressed genes according to immune score (A) and stromal score strata (B).
(C) A dendrogram of clustered genes in WGCNA. Each branch in the figure represents a gene and each color lump below represents a co-expressed gene module.
(D) Correlation heatmap showing the relationship between the gene modules and immune/stromal scores. Robust TME-related gene modules are marked with black frames. The number within the color lump indicates the correlation coefficient, followed by a statistical p value in the parenthesis.
(E) Screening of candidate model genes. The Venn diagram shows the intersection genes between DEGs and robust TME-related gene modules identified by WGCNA. These overlapping genes are then inputted into a LASSO regression analysis. The vertical dashed lines in the middle panel represent the selected appropriate lambda value. Immune score-related and stromal score-related LASSO output genes are combined to generate candidate model genes. Genes in bold in the rectangular boxes are unique genes in immune-related/stromal-related genes. DEGs, differentially expressed genes; WGCNA, weighted gene co-expression network analysis; LASSO, Least Absolute Shrinkage and Selection Operator. See also Figures S1–S4.