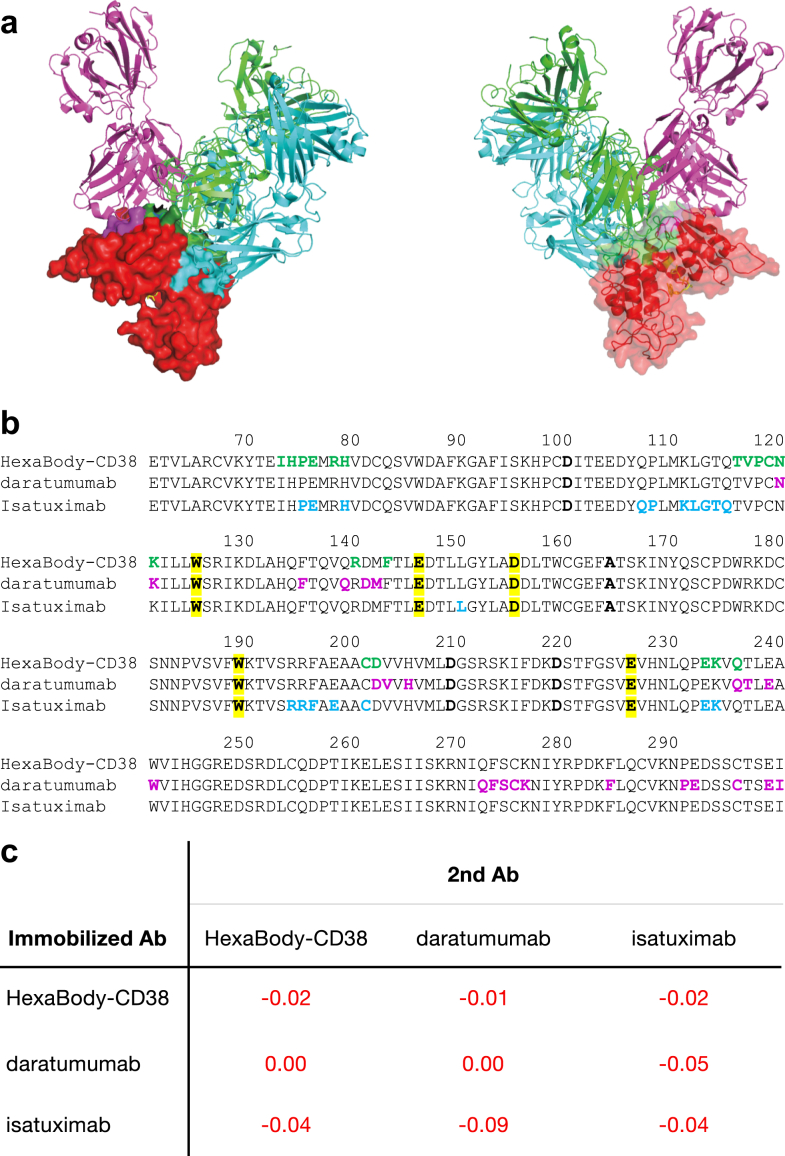
Fig. 6.
Binding competition between HexaBody-CD38, daratumumab, and isatuximab to CD38. HexaBody-CD38 recognizes a unique epitope on CD38 that is distinct from daratumumab and isatuximab. (a) Overlayed co-crystal structures of the Fab-arms of HexaBody-CD38 (green cartoon), daratumumab (7DHA24; magenta cartoon), and isatuximab (4CMH21; cyan cartoon) with soluble CD38 (red surface, transparent in left panel), aligned on the CD38 structures shown from the front and back (left and right panel). CD38 residues that are within a 3.9 Å contact radius with the Fabs are indicated on the surface of CD38 in green, magenta and cyan for HexaBody-CD38, daratumumab and isatuximab, respectively, whereas residues that interact with more than one Fab are shown in dark green. The catalytic site is indicated by a substrate analogue in yellow, superimposed from a published complex of CD38 with PDB code 3P5S.25 (b) Primary amino acid sequence of CD38, numbered according to Uniprot accession P28907-1, with contact residues coloured the same as in (a). Catalytic residues20 are highlighted in yellow. Four point mutants that were introduced to remove glycosylation sites are indicated in bold and are not contact residues in the complexes. (c) CD38 crossblock analysis for HexaBody-CD38, daratumumab, and isatuximab was performed by BLI assay with recombinant human CD38. The numbers in the matrix indicate the response of the second antibody (nm). All responses were <0.1 nm and were considered to be blocking pairs.