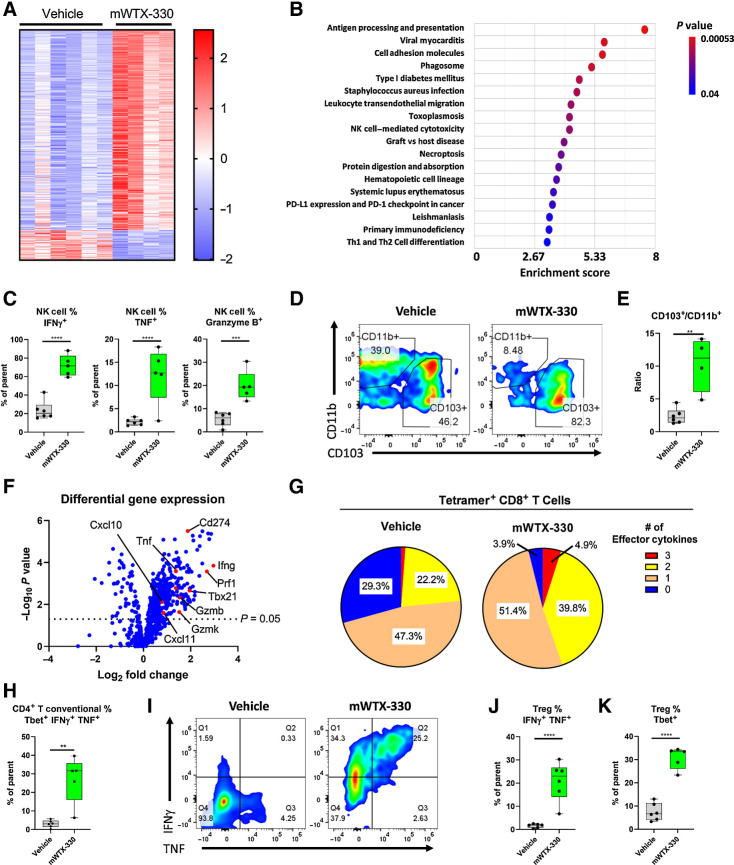
Figure 3.
mWTX-330 activates various tumor-infiltrating immune-cell populations in the MC38 syngeneic tumor model. MC38 tumor-bearing mice were dosed twice a week with mWTX-330 or vehicle. Tumors (n = 5–6 mice per condition) were collected 24 hours after the second dose and dissociated into single cell suspensions for further analysis. RNA from each tumor was isolated and subjected to immune profiling with the NanoString PanCancer Mouse Immune Profiling Panel. A, Heat map of transcripts with statistically significant differences in expression between the two treatments (Supplementary Table S2). Transcripts were excluded from the heat map if they had average normalized counts below 50. Each lane represents an individual animal. B, Pathway enrichment analysis was performed using Partek software. C, The frequency of tumor-infiltrating NK cells producing IFNγ, TNF, or Granzyme B. Representative flow plots of CD11b+ and CD103+ tumor-infiltrating DCs (D) and the resulting ratio (E). F, Volcano plot of transcripts differentially expressed between mWTX-330 and vehicle-treated mice (Supplementary Table S2). G, The frequency of polyfunctional, tetramer positive CD8+ T cells was measured by examining coexpression of IFNγ, TNF, and Granzyme B after PMA/ionomycin restimulation. H, The frequency of conventional CD4+ T cells with a TH1 phenotype (Tbet+IFNγ+TNF+). I, The frequency of tumor-infiltrating FoxP3+ Tregs producing IFNγ and TNF (I–J) or expressing Tbet (K). Unless otherwise stated, data are presented a box and whiskers plot, where the box represents the 25th to 75th percentile, and the middle line represents the mean, whereas P values are derived from t tests. **, P < 0.01; ***, P < 0.001; ****, P < 0.0001.