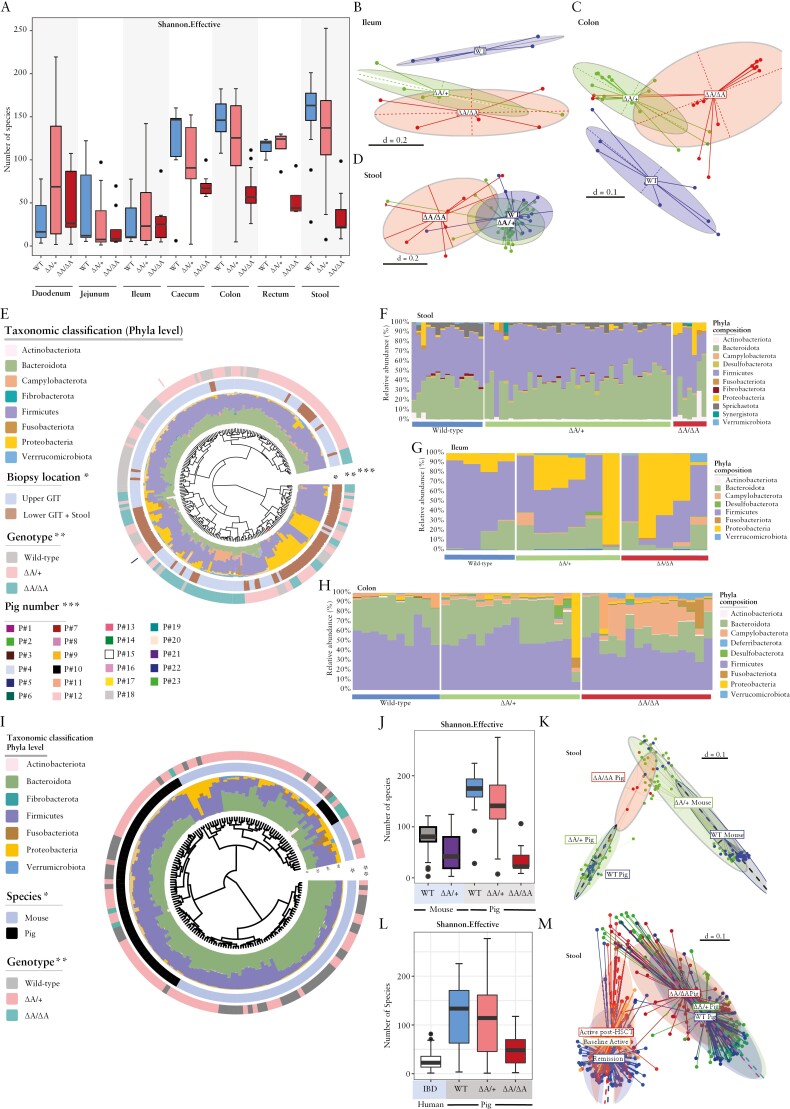
Figure 3.
[A] Alpha-diversity of the luminal and mucosa-associated microbiota. Shannon an effective number of species. Sample locations and genotypes are indicated. [B–D] Multidimensional scaling [MDS] plots of microbial profiles of faecal, ileal and colonic samples stratified by genotype, respectively. [E] Phylogenetic tree showing the similarities between microbiota profiles based on generalized UniFrac distances in luminal [n = 61] and mucosa-associated microbiota [n = 166] derived from 23 pigs [seven wild-type, 16 TNFΔARE]. Individual taxonomic composition at the phylum level is shown as stacked bar plots around the phylogram. Innermost ring shows stratification based on sample type, upper GI tract samples [blue] and lower GI tract and stool samples [brown] and denoted by an asterisk [*]; the second ring shows stratification based on genotype, wild-type [grey], TNFΔARE/+ [pink] and TNFΔARE/ΔARE [green] and denoted by two asterisks [**]. Bars in the outer ring of the figure indicate samples derived from each pig and denoted by three asterisks [***]. [F–H] Taxonomic composition at the phylum level in faecal, ileal and colonic samples stratified by genotype. [I] Phylogenetic tree showing the similarities between microbiota profiles based on generalized UniFrac distances in luminal microbiota derived from wild-type and TNFΔARE pigs and mice. Individual taxonomic composition at the phylum level is shown as stacked bar plots around the phylogram. Innermost ring shows stratification based on species, mouse or pig; and the outer ring shows stratification based on genotype, wild-type [grey], TNFΔARE/+ [pink] and TNFΔARE/ΔARE [green]. [J] Alpha-diversity of luminal microbiota from wild-type and TNFΔARE pigs and mice is shown as Shannon effective number of species. [K] MDS plot of microbial profiles of faecal samples stratified by species [mouse or pig] and genotype, respectively. [L] Alpha-diversity of luminal and mucosal microbiota from human IBD patients and wild-type and TNFΔARE pigs is shown as Shannon effective number of species. [M] MDS plot of microbial profiles of samples stratified by species [human or pig] and disease activity or genotype, respectively.