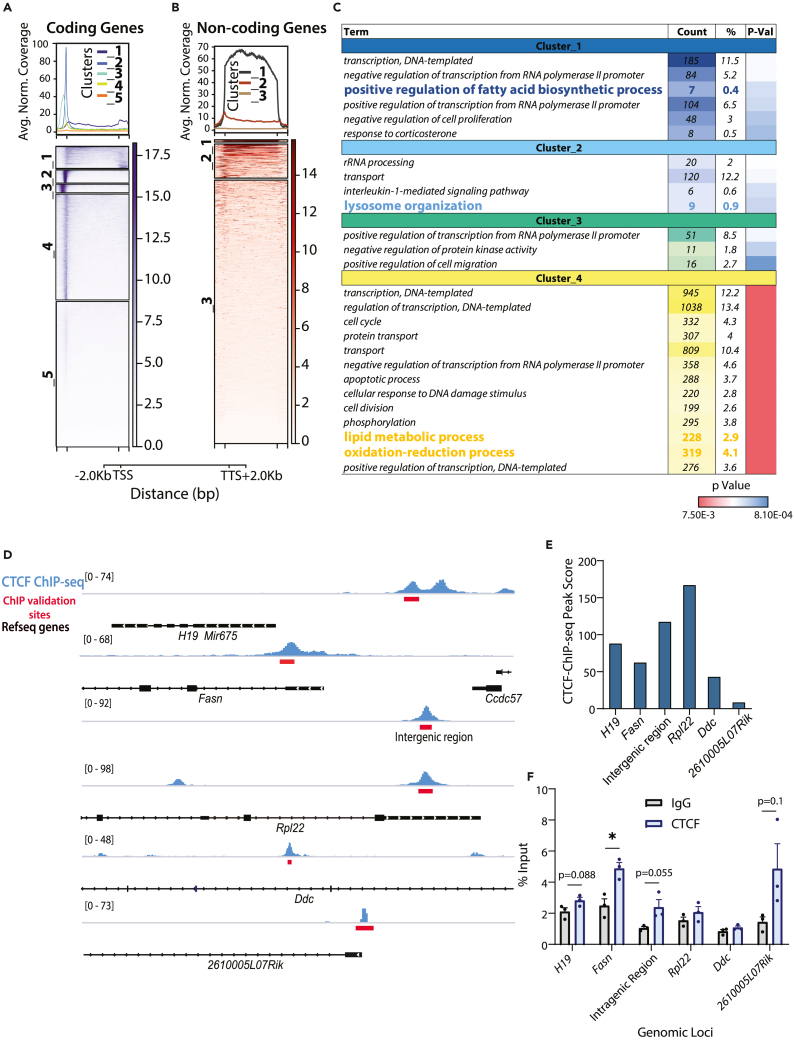
Figure 2.
CTCF occupancy around protein-coding and non-coding genes
(A and B) k-means clustering of genes ((A) protein-coding and (B) non-coding) based on CTCF ChIP-seq signal intensity profile averaged from n = 3 (across gene body and 2Kb upstream and downstream of TSS and TTS, respectively).
(C) GO-BP (Biological Process) terms enriched in genes associated with cluster 1–4 (protein-coding genes).
(D) Genome Browser (IGV) snapshots of CTCF binding intensities on genes as indicated. Red boxes depict genomic locus where CTCF binding has been validated using ChIP-qPCR.
(E and F) CTCF-ChIP-seq peaks scores (E) and ChIP-qPCR validation (n = 3, representative from N = 2) (F) of CTCF binding at genomic loci shown in D. Data are represented as mean ± SEM. For F, statistical significance was calculated by using a Student’s t test (∗, p < 0.05; ∗∗, p < 0.01; ∗∗∗, p < 0.001). TSS-Transcription Start Site, TTS-Transcription Termination Site.