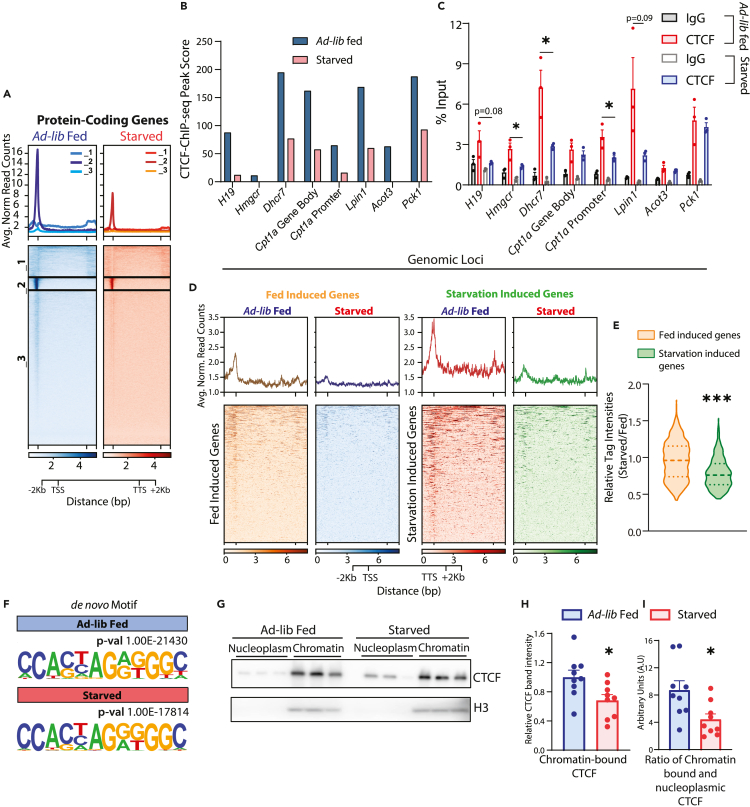
Figure 5.
Starvation-induced reduction in genome-wide chromatin occupancy of CTCF
(A) Profile of CTCF ChIP-seq signal intensities (normalized using respective input controls) on protein-coding genes in ad-lib fed and 24 h starved livers and clustered (k-mean) based on CTCF binding profile across gene body and 2 kb upstream and downstream of TTS and TTS, respectively.
(B and C) CTCF-ChIP-seq peaks scores (B) and ChIP-qPCR validation (n = 3, representative from N = 2) (C) in ad-lib fed (11 p.m.) and 24 h starved (8 p.m. onwards) mice livers at the CTCF binding sites shown in Figure S4E (Green Boxes).
(D) Profile of CTCF-ChIP-seq signal intensities in ad-lib fed and 24 h starved liver on genes that are differentially expressed in respective conditions (Figure 3A).
(E) Relative CTCF ChIP-seq signal intensity (24 h starved/ad-lib fed) at promoter (TSS ±3 kb) of feeding and starvation-induced genes (n = 3).
(F) De novo motifs enriched at CTCF peaks in ad-lib fed and 24 h starved liver.
(G) Representative western blot of chromatin-associated and nucleoplasmic CTCF in ad-lib fed (11 p.m.) and 24 h starved (8 p.m. onwards) mice livers.
(H and I) Quantifications of (H) CTCF band intensities in chromatin fraction and (I) Ratio of chromatin-associated and nucleoplasmic CTCF in ad-lib fed (11 p.m.) and 24 h of starvation (8 p.m. onwards) mice livers. In H, CTCF band intensities from the chromatin fraction were normalized using H3 and for I, it has been further divided by the CTCF band intensities from the nucleoplasmic fraction (N = 3, n = 3). Data are represented as mean ± SEM. For C, E, H, & I statistical significance was calculated by using a Student’s t test (∗, p < 0.05; ∗∗, p < 0.01; ∗∗∗, p < 0.001). TSS-Transcription Start Site, TTS-Transcription Termination Site.