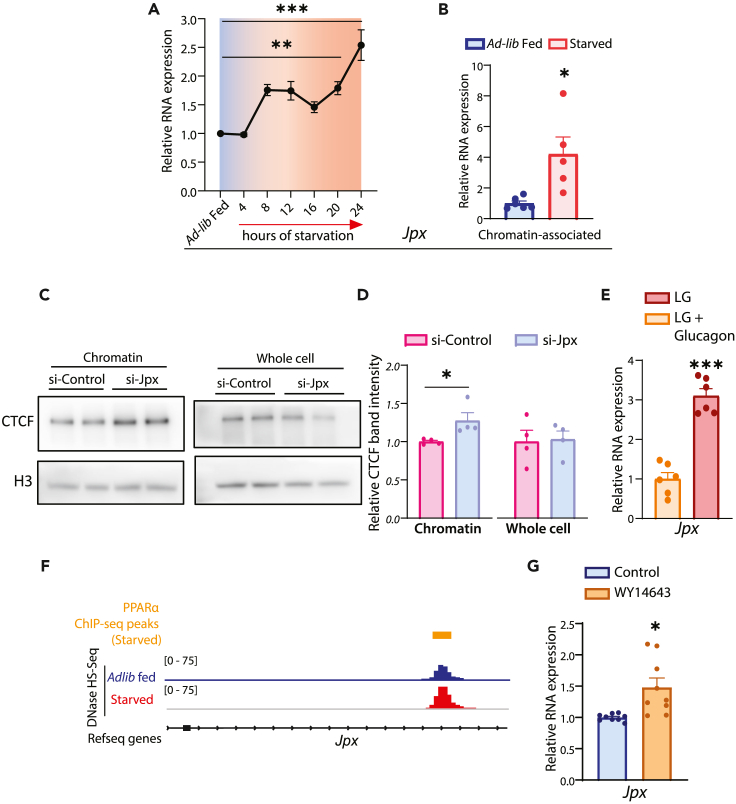
Figure 6.
Starvation-dependent induction of Jpx expression in the liver and its effects on CTCF chromatin occupancy
(A) Relative RNA expression of Jpx in ad-lib fed (7 a.m.) and after different duration (4, 8, 12, 16, 20 and 24 h) of starvation (7 a.m. onwards) in mice liver. Jpx RNA was normalized to actin (Actb) transcript levels and plotted as fold change with respect to ad-lib fed (N = 2, n = 3).
(B) Relative expression of chromatin-bound Jpx in ad-lib fed and 24 h starved livers, normalized to Hotair transcript levels and plotted as fold change with respect to ad-lib fed (N = 2, n = 2–3).
(C) Representative immunoblot of chromatin-associated (Left) and whole cell (Right) CTCF in primary hepatocytes transfected with si-Jpx (Jpx KD)/si-Control (Control KD).
(D) Quantification of chromatin-associated CTCF in primary hepatocytes transfected with si-Jpx/si-Control. CTCF band intensities were quantified with H3 as a loading control and presented as fold change in comparison to si-Control (N = 2, n = 2).
(E) Relative RNA expression of Jpx in primary hepatocytes treated with glucagon (30 nM) in LG medium for 3 h, normalized with actin (Actb) transcript levels and plotted as fold change with respect to untreated cells (N = 2, n = 3).
(F) Genome Browser (IGV) snapshots of PPARα ChIP-seq peaks in the starved liver and DNase I Hypersensitivity-sites (DHS-seq) in ad-lib fed and 24 h starved liver at Jpx gene-coding locus.
(G) Relative RNA expression of Jpx in primary hepatocytes treated with WY14643 (50 μM) in HG media for 24 h, normalized with actin (Actb) transcript levels and plotted as fold change with respect to untreated cells (N = 2, n = 3). Data are represented as mean ± SEM. For A, statistical significance was calculated by using one-way ANOVA with Dunnett’s test for multiple comparison against ad-lib fed control and for B, D, E & G, using a Student’s t test (∗, p < 0.05; ∗∗, p < 0.01; ∗∗∗, p < 0.001).