Abstract
An assessment was conducted on the level of inactivation of relevant pathogens that could be present in processed animal protein of porcine origin intended to feed poultry and aquaculture animals when methods 2 to 5 and method 7, as detailed in Regulation (EU) No 142/2011, are applied. Five approved scenarios were selected for method 7. Salmonella Senftenberg, Enterococcus faecalis, spores of Clostridium perfringens and parvoviruses were shortlisted as target indicators. Inactivation parameters for these indicators were extracted from extensive literature search and a recent EFSA scientific opinion. An adapted Bigelow model was fitted to retrieved data to estimate the probability that methods 2 to 5, in coincidental and consecutive modes, and the five scenarios of method 7 are able to achieve a 5 log10 and a 3 log10 reduction of bacterial indicators and parvoviruses, respectively. Spores of C. perfringens were the indicator with the lowest probability of achieving the target reduction by methods 2 to 5, in coincidental and consecutive mode, and by the five considered scenarios of method 7. An expert knowledge elicitation was conducted to estimate the certainty of achieving a 5 log10 reduction of spores of C. perfringens considering the results of the model and additional evidence. A 5 log10 reduction of C. perfringens spores was judged: 99–100% certain for methods 2 and 3 in coincidental mode; 98–100% certain for method 7 scenario 3; 80–99% certain for method 5 in coincidental mode; 66–100% certain for method 4 in coincidental mode and for method 7 scenarios 4 and 5; 25–75% certain for method 7 scenario 2; and 0–5% certain for method 7 scenario 1. Higher certainty is expected for methods 2 to 5 in consecutive mode compared to coincidental mode.
Keywords: animal by‐products, porcine, inactivation, pathogens, perfringens, feed
Summary
Following the partial revision of the feed ban introduced by Commission Regulation (EU) 2021/1372 the European animal by‐product processing sector (EFPRA) asked the Commission to revise the standards for the production of processed animal protein (PAP) of porcine origin set out in Regulation (EU) No 142/2011. In accordance with specific requirements set out in this Regulation, to be placed on the market, PAP of mammalian origin intended to feed poultry and aquaculture animals must have been submitted to processing method 1 (pressure sterilisation) as set out in Chapter III of Annex IV to Regulation (EU) No 142/2011. However, the rendering industry produces PAP intended for pet food, fertilisers or fuel for combustion applying primarily methods 2 to 5 or method 7 as set out in the Regulation. Therefore, the Commission requested EFSA to provide a scientific opinion concerning the efficacy of methods 2 to 5 and method 7 to inactivate relevant pathogens when producing PAP of porcine origin intended to feed poultry and aquaculture animals. In particular, the assessment concerned the level of inactivation of relevant pathogens that could be present in PAP of porcine origin intended to feed poultry and aquaculture animals.
The Term of Reference was translated into five assessment questions (AQ). AQ1: what relevant pathogens can be used as indicators to assess the efficacy of standard processing methods for Category 3 ABP of porcine origin? AQ2: what are the technical parameters (e.g. time, temperature, pressure, pH, particle size) for methods 2 to 5 and 7? AQ3: what are the inactivation parameters (D values, z) of the relevant pathogens identified in AQ1? AQ4: what is the ‘level of inactivation’ of the selected relevant pathogens achieved by methods 2 to 5 and 7? AQ5: what is the certainty that the ‘level of inactivation’ achieved by methods 2 to 5 and 7, as in AQ4, is sufficient to reach the standards for Category 3 ABP of porcine origin? The approach to answer the ToR is described in the protocol (Annex A).
To identify what relevant pathogens (AQ1) (i.e. bacteria, parasites and viruses) can be used as indicators to assess the efficacy of the methods under assessment, a set of criteria were developed based on previous EFSA standards and scientific opinions, the EU legislation, the WOAH list of swine diseases and an extensive literature search (for viruses only). As a result, Salmonella Senftenberg, Enterococcus faecalis and spores of Clostridium perfringens were selected as relevant bacterial pathogens in PAP of porcine origin. Relevant viral hazards in PAP of porcine origin identified were: porcine adenovirus (Adenoviridae), Torque Teno virus (Anelloviridae), porcine circovirus (Circoviridae), bocavirus and porcine parvovirus (Parvoviridae). Taenia solium and Trichinella spp were considered relevant parasitic pathogens in PAP of porcine origin.
The technical parameters of methods 2 to 5 (AQ2) are stated in Chapter III of Annex IV to Regulation (EU) No 142/2011: method 2, 100°C for at least 125 min, 110°C for at least 120 min and 120°C for at least 50 min; method 3, 100°C for at least 95 min, 110°C for at least 55 min and 120°C for at least 13 min; method 4, 100°C for at least 16 min, 110°C for at least 13 min, 120°C for at least 8 min and 130°C for at least 3 min; method 5, 80°C for at least 120 min and 100°C for at least 60 min. The Commission Regulation (EU) 142/2011 specifies that the core temperatures specified above may be achieved consecutively or through a coincidental combination of the time periods indicated. The coincidental mode was considered the worst‐case scenario and selected for the assessment of the efficacy of methods 2 to 5 to inactivate relevant pathogens. For method 7, the legislation does not state time/temperature parameters. However, there are process parameters approved in different EU member states based on equipment used and treated raw materials. Based on the approved parameters provided by the industry, five scenarios of individual time/temperature profiles were selected for this assessment in order to apply for method 7 the same methodological approach than for methods 2 to 5: scenario 1, 80°C for at least 14 min; scenario 2, 95°C for at least 90 min; scenario 3, 115°C for at least 56 min; scenario 4, 125°C for at least 10 min; scenario 5, 133°C for at least 5 min.
The thermal inactivation parameters (AQ3) of spores of C. perfringens were retrieved through an extensive literature search (ELS), while those of S. Senftenberg and E. faecalis were collated in a previous EFSA scientific opinion (EFSA BIOHAZ Panel, 2021). The thermal inactivation parameters of the relevant viral families were retrieved through an ELS and from a previous EFSA scientific opinion (EFSA BIOHAZ Panel, 2021). Parvoviruses were selected as the most thermal resistant viral hazards for the assessment. For the parasites, the thermal resistance parameters were not retrieved because the thermal resistance of parasites is lower than that of the relevant bacterial and viral pathogens.
An adapted Bigelow model was fitted to the thermal inactivation parameters retrieved for the selected pathogens to estimate the probability that the time/temperature combinations of methods 2 to 5 as stated in the regulation, in both coincidental and consecutive mode, and of the five scenarios of method 7, set for this assessment, are able to achieve a 5 log10 reduction of bacterial indicators and a 3 log10 reduction of parvoviruses (AQ4). C. perfringens was the indicator with the lowest probability of achieving the target 5 log10 reduction by methods 2, 3, 4, 5 in coincidental mode and the five time/temperature combinations of method 7. The results of the model showed a probability of inactivation of at least 5 log10 of spores of C. perfringens of over 0.99 for methods 2, 3 and 5, in both the coincidental and consecutive modes and of 0.92 for method 4 in consecutive mode. For method 4 in coincidental mode, the model estimated a probability of 0.066. For method 7, the model estimated a probability of 0.004, or below, of achieving the 5 log10 reduction for scenarios 1, 4 and 5, and probabilities of 0.685 and 0.999 of achieving the same level of reduction for scenarios 2 and 3, respectively. For method 4, in coincidental mode, and scenarios 4 and 5 of method 7, the low probabilities are possibly associated with the fact that, when modelling, no extrapolation at temperatures other than those with experimental data available (105°C for spores of C. perfringens) was applied while the processing methods involved much higher temperatures (in the range 125–133°C). This would lead to an underestimation of the level of inactivation reached.
An expert knowledge elicitation (EKE) was conducted to elucidate what the probability is that a 5 log10 reduction of C. perfringens spores is achieved, in more than 99% of cases, by application of the relevant processes (methods 2, 3, 4, 5 in coincidental mode and five t/T combinations of method 7) assuming that the processes are performed as prescribed and that the indicated process conditions are achieved (AQ5). Based on the EKE, the certainty of achieving a 5 log10 reduction of C. perfringens spores (which also assure the target inactivation for the other relevant pathogens) was: 99–100% certain for methods 2 and 3 in coincidental mode; 98–100% certain for method 7 scenario 3; 80–99% certain for method 5 in coincidental mode; 66–100% certain for method 4 in coincidental mode and for method 7 scenarios 4 and 5; 25–75% certain for method 7 scenario 2; and 0–5% certain for method 7 scenario 1. Compared to the results of the EKE for methods 2–5 in coincidental mode, the same or higher certainty to achieve the 5 log10 reduction of C. perfringens spores is expected when methods 2 to 5 are applied in consecutive mode.
1. Introduction
1.1. Background and Terms of Reference as provided by the requestor
Following the partial revision of the feed ban introduced by Commission Regulation (EU) 2021/1372 1 the European animal by‐product processing sector (EFPRA) asked the Commission to revise standards for the production of processed animal protein (PAP) of porcine origin set out in Point B (1) of Section 1 of Chapter II of Annex X to Regulation (EU) No 142/2011 2 .
PAP is defined in point 5 of Annex I of Regulation (EU) No 142/2011 as Category 3 materials referred to in Article 10 of Regulation (EC) No 1069/2009 3 . PAP must comply with general requirements for the processing and placing on the market set out in Chapter I Annex X of Regulation (EU) No 142/2011. In accordance with specific requirements for processing of PAP set out in Point B (1) of Chapter II of that Annex, PAP of mammalian origin must have been submitted to processing method 1 (pressure sterilisation) as set out in Chapter III of Annex IV to Regulation (EU) No 142/2011.
By way of derogation set out in Point B(1)(b) PAP of mammalian origin may have been submitted to any of the processing methods 1 to 5 or processing method 7, as set out in Chapter III of Annex IV to Regulation (EU) No 142/2011 when disposed of or used as a fuel for combustion or use in petfood.
In accordance with point 1(b) of Section 1 of Chapter II of Annex XI to Regulation (EU) No 142/2011, the above derogation is applicable also to PAP intended for the manufacturing of organic fertilizers and soil improvers (fertilizers).
Before publication of Commission Regulation (EU) 2021/1372, the use of PAP of mammalian origin was limited to the manufacturing of petfood, feed for aquaculture animals (since 2013), and fertilizers or as fuel for combustion, as described above.
After the partial revision of the feed ban, PAP of porcine origin may be also fed to poultry. However, following decades of a quasi‐complete feed ban on protein of terrestrial animal's origin, rendering industry produces only PAP intended for petfood, fertilizers or fuel for combustion applying methods 2 to 5 or method 7. According to the information from EFPRA, currently no EU operator applies method 1 for the processing of PAP of porcine origin.
The Commission would therefore like to explore the efficacy of methods 2 to 5 or method 7 to inactivate relevant pathogens when producing PAP of porcine origin intended to feed poultry and aquaculture animals.
Terms of Reference (ToR)
In the light of the above, and in accordance with Article 29 of Regulation (EC) No 178/2002 4 , the Commission requests EFSA to provide a scientific opinion concerning the efficacy of methods 2 to 5 and method 7 to inactivate relevant pathogens when producing processed animal protein (PAP) of porcine origin intended to feed poultry and aquaculture animals.
In particular, the scientific opinion should comprise an assessment of the level of inactivation of relevant pathogens that could be present in processed animal protein of porcine origin intended to feed poultry and aquaculture animals.
1.2. Interpretation of the Terms of Reference
Initial clarification was requested to the European Commission on whether the Category 3 material considered by the mandate should be only of EU origin and not sourced from third countries, since the criteria to select relevant pathogens could differ. Subsequently, since the WG decided to work with indicators (see AQ1 of the protocol in Annex A) and the EC agreed with the approach (see Section 3.2.1), the origin of the Category 3 material was not considered relevant. No further clarification was requested on the source of the materials nor on the criteria for selection of relevant pathogens.
The ToR has been translated into assessment questions (AQ) and sub‐questions (SQ), as follows:
AQ1: What relevant pathogens can be used as indicators to assess the efficacy of standard processing methods as in Chapter III of Annex IV to Regulation (EU) No 142/2011, for Category 3 ABP of porcine origin?
AQ2: What are the technical parameters (e.g. time, temperature, pressure, pH, particle size) for methods 2 to 5 and 7 as in Chapter III of Annex IV to Regulation (EU) No 142/2011?
SQ2.1: What are the technical parameters (e.g. time, temperature, pressure, pH, particle size) for methods 2, 3, 4 and 5 as in Chapter III of Annex IV to Regulation (EU) No 142/2011?
SQ2.2: What are the technical parameters for methods 7 approved at national level in the EU?
AQ3: What are the inactivation parameters (D and z) of the relevant pathogens identified in AQ1?
AQ4: What is the ‘level of inactivation’ of the selected relevant pathogens achieved by methods 2 to 5 and 7?
SQ4.1 What is the ‘level of inactivation’ achieved by methods 2, 3, 4 and 5?
SQ4.2 What is the ‘level of inactivation’ achieved for method 7?
AQ5: What is the certainty that the ‘level of inactivation’ achieved by methods 2, 3, 4, 5 and 7, as in AQ4, is sufficient to reach the standards for Category 3 ABP of porcine origin?
Further processing of porcine PAP before they are included in feed for poultry and aquaculture may contribute to further reduction of relevant pathogens. These subsequent processing steps have not been considered in this assessment.
Any assessments conducted by the competent authorities to approve applications of method 7 have not been considered in this opinion, precluding any conclusion about the validity or appropriateness of such assessments.
1.3. Additional information (if appropriate)
1.3.1. Technical parameters (time, temperature, pressure, pH, particle size) of methods 2 to 5 and 7 (AQ2)
Chapter III, Annex IV of Commission Regulation (EU) 142/2011 describes the standard processing methods of ABP, summarised in Figure 1, as follows (A. Processing method 1 (pressure sterilisation) and F. processing method 6 are not described):
Figure 1.
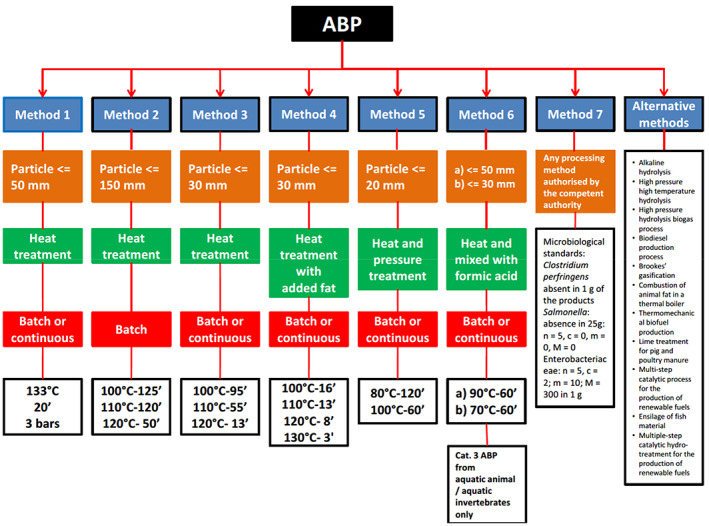
Summary of the processing methods of ABP according to Commission Regulation (EU) No 142/2011. Source: adapted from the Spanish Renderers Association (ANAGRASA) website: https://www.anagrasa.org/es/sector/preguntas-frecuentes/index.htm
B. Processing method 2
Reduction
If the particle size of the animal by‐products to be processed is more than 150 millimetres, the animal by‐products must be reduced in size using appropriate equipment, set so that the particle size after reduction is no greater than 150 millimetres. The effectiveness of the equipment must be checked daily and its condition recorded. If checks disclose the existence of particles larger than 150 millimetres, the process must be stopped and repairs made before the process is resumed.
Time, temperature and pressure
-
2
After reduction the animal by‐products must be heated in a manner, which ensures that a core temperature greater than 100°C is achieved for at least 125 min, a core temperature greater than 110°C is achieved for at least 120 min and a core temperature greater than 120°C is achieved for at least 50 min. The core temperatures may be achieved consecutively or through a coincidental combination of the time periods indicated.
-
3
The processing must be carried out in a batch system.
C. Processing method 3
Reduction
If the particle size of the animal by‐products to be processed is more than 30 millimetres, the animal by‐products must be reduced in size using appropriate equipment, set so that the particle size after reduction is no greater than 30 millimetres. The effectiveness of the equipment must be checked daily and its condition recorded. If checks disclose the existence of particles larger than 30 millimetres, the process must be stopped and repairs made before the process is resumed.
Time, temperature and pressure
-
2
After reduction the animal by‐products must be heated in a manner, which ensures that a core temperature greater than 100°C is achieved for at least 95 min, a core temperature greater than 110°C is achieved for at least 55 min and a core temperature greater that 120°C is achieved for at least 13 minutes. The core temperatures may be achieved consecutively or through a coincidental combination of the time periods indicated.
-
3
The processing may be carried out in batch or continuous systems.
D. Processing method 4
Reduction
If the particle size of the animal by‐products to be processed is more than 30 millimetres, the animal by‐products must be reduced in size using appropriate equipment, set so that the particle size after reduction is no greater than 30 millimetres. The effectiveness of the equipment must be checked daily and its condition recorded. If checks disclose the existence of particles larger than 30 millimetres, the process must be stopped and repairs made before the process is resumed.
Time, temperature and pressure
-
2
After reduction the animal by‐products must be placed in a vessel with added fat and heated in a manner which ensures that a core temperature greater than 100°C is achieved for at least 16 min, a core temperature greater than 110°C is achieved for at least 13 min, a core temperature greater than 120°C is achieved for at least 8 min and a core temperature greater that 130°C is achieved for at least 3 min. The core temperatures may be achieved consecutively or through a coincidental combination of the time periods indicated.
-
3
The processing may be carried out in batch or continuous systems.
E. Processing method 5
Reduction
If the particle size of the animal by‐products to be processed is more than 20 millimetres, the animal by‐products must be reduced in size using appropriate equipment, set so that the particle size after reduction is no greater than 20 millimetres. The effectiveness of the equipment must be checked daily and its condition recorded. If checks disclose the existence of particles larger than 20 millimetres, the process must be stopped and repairs made before the process is resumed.
Time, temperature and pressure
-
2
After reduction the animal by‐products must be heated until they coagulate and then pressed so that fat and water are removed from the proteinaceous material. The proteinaceous material must then be heated in a manner which ensures that a core temperature greater than 80°C is achieved for at least 120 min and a core temperature greater that 100°C is achieved for at least 60 min.
The core temperatures may be achieved consecutively or through a coincidental combination of the time periods indicated.
-
3
The processing may be carried out in batch or continuous systems.
G. Processing method 7
- Any processing method authorised by the competent authority where the following have been demonstrated by the operator to that authority:
- the identification of relevant hazards in the starting material, in view of the origin of the material, and of the potential risks in view of the animal health status of the Member State or the area or zone where the method is to be used;
- the capacity of the processing method to reduce those hazards to a level, which does not pose any significant risks to public and animal health;
- the sampling of the final product on a daily basis over a period of 30 production days in compliance with the following microbiological standards:
-
Samples of material taken directly after the treatment:Clostridium perfringens absent in 1 g of the products.
-
Samples of material taken during or upon withdrawal from storage:Salmonella: absence in 25 g: n = 5, c = 0, m = 0, M = 0.Enterobacteriaceae: n = 5, c = 2; m = 10; M = 300 in 1 g.where:n = number of samples to be tested;m = threshold value for the number of bacteria; the result is considered satisfactory if the number of bacteria in all samples does not exceed m;M = maximum value for the number of bacteria; the result is considered unsatisfactory if the number of bacteria in one or more samples is M or more; and;c = number of samples the bacterial count of which may be between m and M, the samples still being considered acceptable if the bacterial count of the other samples is m or less.
-
Details of the critical control points under which each processing plant satisfactorily complies with the microbiological standards must be recorded and maintained so that the operator and the competent authority can monitor the operation of the processing plant. The information to be recorded and monitored must include the particle size, and, as appropriate, the critical temperature, the absolute time, pressure profile, raw material feed rate and fat recycling rate.
By way of derogation from point 1, the competent authority may authorise the use of processing methods which have been approved prior to the date of entry into application of this Regulation, in accordance with Chapter III of Annex V to Regulation (EC) No 1774/2002.
The competent authority shall permanently or temporarily suspend the application of processing methods referred to in points 1 and 3, if it obtains evidence that any of the circumstances specified in point 1(a) or (b) have substantially changed.
The competent authority shall inform the competent authority of another Member State upon request about the information at its disposal under points 1 and 2 in relation to an authorised processing method.
1.3.2. Authorised method 7 processes
The European Fat Processors and Renderers Association (EFPRA) shared with EFSA a total of 21 process parameters approved in the EU since 2004 for method 7 to support the assessment to answer the ToR of this mandate.
The provided parameters refer to both in‐continuous and in‐batch processes, with temperatures ranging between 85°C and 133°C and lasting between 2 and 255 min. The particle sizes of the processed materials range between < 20 and 50 mm and the maximum pressure applied is 3.5 bars. The raw materials for which method 7 has been approved include blood, hair and melted fat and originate from different animal species, including pigs. The process parameters were shared with EFSA confidentially and the WG selected five time/temperature scenarios out of them (see Section 2.3.1). The scenarios were selected to include the widest range of temperatures among those reported using starting material of porcine or mixed origin, in continuous and batch modes.
1.3.3. Processed animal proteins and feed
Processed Animal Protein (PAP) is defined in Annex I of Commission Regulation (EU) No 142/2011, as animal protein derived entirely from Category 3 material, which have been treated in accordance with Section 1 of Chapter II of Annex X (including blood meal and fishmeal) so as to render them suitable for direct use as feed material or for any other use in feedingstuffs, including petfood, or for use in organic fertilisers or soil improvers; however, it does not include blood products, milk, milk‐based products, milk‐derived products, colostrum, colostrum products, centrifuge or separator sludge, gelatine, hydrolysed proteins and dicalcium phosphate, eggs and egg‐products, including eggshells, tricalcium phosphate and collagen.
According to Article 31 of Regulation (EC) No 1069/2009, animal by‐products and derived products destined for feeding to farmed animals, excluding fur animals, may only be placed on the market provided:
- they are derived from Category 3 material other than:
- hides and skins, hooves, feathers, wool, horns, hair and fur originating from dead animals that did not show any signs of disease communicable through that product to humans or animals (Article 10 (n) Regulation (EC) No 1069/2009)
- adipose tissue from animals which did not show any signs of disease communicable through that material to humans or animals, which were slaughtered in a slaughterhouse, and which were considered fit for slaughter for human consumption following an ante‐mortem inspection in accordance with Community legislation (Article 10 (o) Regulation (EC) No 1069/2009)
- catering waste (Article 10 (p) Regulation (EC) No 1069/2009)
they have been collected or processed, as applicable, in accordance with the conditions for pressure sterilisation or other conditions to prevent risks arising to public and animal health in accordance with measures adopted pursuant to Article 15 of Regulation (EC) No 1069/2009 and any measures which have been laid down in accordance with paragraph 2 of Article 15; and
they come from approved or registered establishments or plants, as applicable for the animal by‐product or derived product concerned.
According to Section 1.A, Chapter II of Annex X of Commission Regulation (EU) No 142/2011 only animal by‐products which are Category 3 material, or products which are derived from such animal by‐products, other than the Category 3 materials referred to in Articles 10 n, o, p of Regulation (EC) No 1069/2009, may be used for the production of PAP.
In point 1.B,
- PAP of mammalian origin must have been submitted to processing method 1 (pressure sterilisation) as set out in Chapter III of Annex IV. However,
- porcine blood or fractions of porcine blood for the production of blood meal may have been submitted instead to any of the processing methods 1 to 5 or processing method 7 as set out in Chapter III of Annex IV, provided that in the case of processing method 7, a heat treatment throughout its substance at a temperature of 80°C has been applied;
- processed animal protein of mammalian origin
- may have been submitted to any of the processing methods 1 to 5 or processing method 7, as set out in Chapter III of Annex IV, provided that it is subsequently disposed of or used as a fuel for combustion;
- where it is exclusively destined for use in petfood, it may have been submitted to any of the processing methods 1 to 5 or processing method 7, as set out in Chapter III of Annex IV, provided that it is
-
–transported in dedicated containers that are not used for the transport of animal by‐products or feedingstuffs for farmed animals, and
-
–consigned directly from a processing plant for Category 3 material to the petfood plant or to an approved storage plant, from where it is directly consigned to a petfood plant
-
–
Non‐mammalian processed animal protein, with the exception of fishmeal, must have been submitted to any of processing methods 1 to 5 or processing method 7, as set out in Chapter III of Annex IV.
Fishmeal must have been submitted to: (a) any of the processing methods set out in Chapter III of Annex IV; or (b) another method which ensures that the product complies with the microbiological standards for derived products set in Chapter I of this Annex.
2. Data and methodologies
The approach to answer the ToR was defined in advance and is described in the protocol (Annex A). Protocol development followed the draft framework for protocol development for EFSA's scientific assessments (EFSA, 2020). It covers both the problem formulation (i.e. what the assessment aims to address) and which methods will be used for addressing the problem. The problem formulation (‘what’) includes the clarification of the mandate (see further refined in Section 1.2) and consists of the steps (1) translation of the mandate into scientifically answerable AQ, (2) definition of the sub‐questions (SQ) of each AQ, if needed, and their relationship (conceptual model) and (3) the selection of the approach for the assessment. The planning of the methods for conducting the assessment (‘how’) consists of (1) specifying the evidence needs and the methods for answering each AQ/SQ, including uncertainty analysis and (2) the methods for integrating evidence across AQ/SQ and addressing the remaining and overall uncertainty.
The methodologies applied for answering some AQ can be fully found in the protocol, while more details are provided below for methods used for other AQ.
2.1. Relevant pathogens that can be used as indicators to assess the efficacy of standard processing methods for Category 3 ABP of porcine origin (AQ1)
An extensive literature search (ELS) was carried out for the identification of viral hazards, which resulted in the extraction of 1,371 records after applying the search strategy described in the protocol (Figure 2). All of them were screened by title and abstract, with 524 being selected for full text screening, and the other 847 excluded because they did not meet the inclusion criteria. Out of the 524 selected for full text screening, and 245 were selected for data extraction. The other 279 were discarded because they were not in English, no full text was available, or they did not meet the eligibility criteria (see protocol in Annex A). Relevant data from the shortlisted 245 references were extracted in tabular format and are presented in Section 3.1.3.
Figure 2.
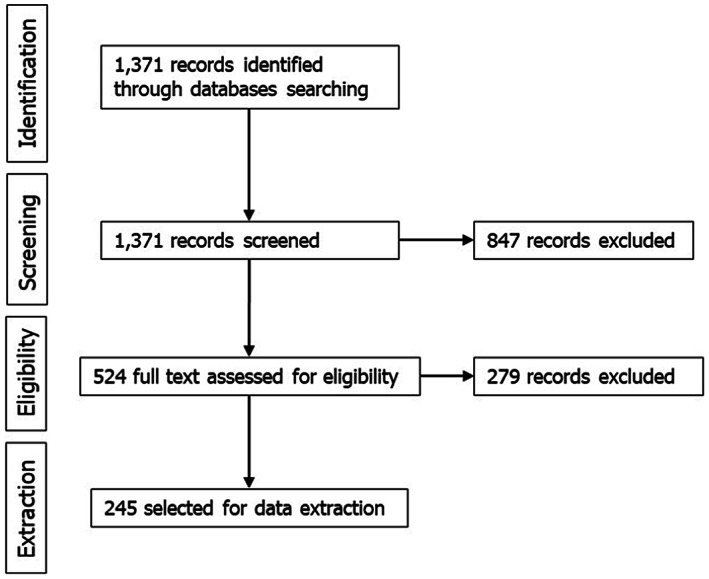
Flow diagram of the literature review for viral hazard identification
2.2. Thermal inactivation parameters (D values) of the relevant biological hazards (AQ3)
An extensive literature search (ELS) was also performed for the identification of thermal inactivation parameters of spores of Clostridium perfringens, which resulted in the extraction of 2,464 records after applying the search strategy described in the protocol (Figure 3). All were screened by title and abstract, with 192 being selected for full text screening, and the other 2,272 excluded because they did not meet the inclusion criteria. Out of the 192 selected for full text screening, 68 were selected for data extraction. The other 124 were discarded because they were not in English, no full text was available, or they did not meet the eligibility criteria (see protocol in Annex A). Relevant data from the shortlisted 68 references were extracted. Of those, D‐values from 18 references were selected for analyses and are described in tabular format in Section 3.2.1.
Figure 3.
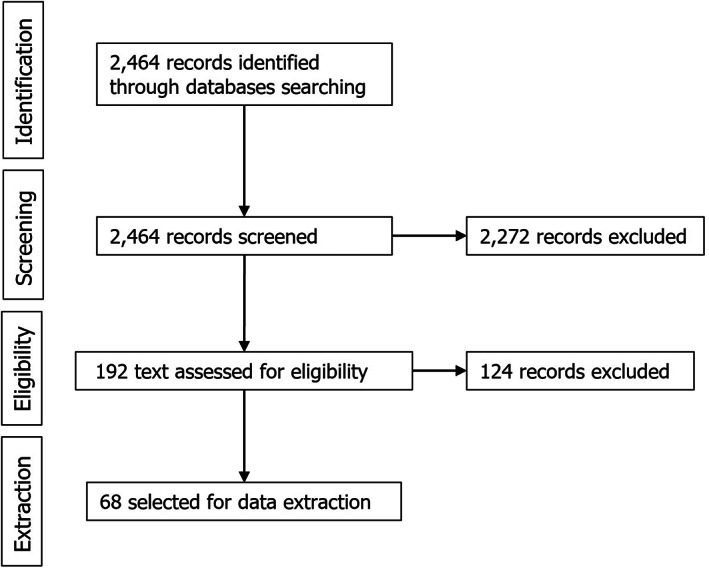
Flow diagram of the literature review for inactivation parameters of Clostridium perfringens
The search string used for the extensive literature search (ELS) for C. perfringens was revised to search for thermal inactivation parameters of the identified relevant viral pathogens (parvoviruses, circoviruses, anelloviruses and adenoviruses), as follows (not included in the protocol):
Title and abstract = [(“thermal inactivat*” OR “heat inactivat*” OR “thermal reduction” OR “heat reduction” OR “thermal survival” OR “heat survival” OR “kill time” OR “thermal kinetic*” OR “heat kinetic*” OR “thermal destruction” OR “heat destruction” OR “thermal process*” OR “thermal treatment*” OR “heat treatment*” OR “thermal resistan*” OR “heat resistan*” OR “thermal performance*” OR “heat performance*” OR “temperature toleran*” OR “heat toleran*” OR “thermal toleran*” OR “time temperature” OR “thermal lethality” OR “heat lethality” OR Bigelow OR “D value*” OR “z value*” OR “F value*” OR “D‐value*” OR “z‐value*” OR “F‐value*” OR “Decimal reduction” OR Sterility OR Pasteuriz* OR Pasteuris* OR Steriliz* OR Sterilis*)] AND [(Parvovirus OR porcine parvovirus OR swine parvovirus OR bocavirus OR human bocavirus OR porcine bocavirus OR swine bocavirus OR parvoviridae OR parvovirinae OR Amdoparvovirus OR Artiparvovirus OR Aveparvovirus OR Bocaparvovirus OR Copiparvovirus OR Dependoparvovirus OR Erythroparvovirus OR Loriparvovirus OR Protoparvovirus OR Tetraparvovirus OR Circovirus OR porcine circovirus OR swine circovirus OR Cyclovirus OR circoviridae OR Torque teno sus virus OR Torque Teno virus OR torqueteno OR TTSuV OR Adenovirus OR porcine adenovirus OR swine adenovirus OR human adenovirus OR HAdV OR Adenoviridae OR Atadenovirus OR Aviadenovirus OR Ichtadenovirus OR Mastadenovirus OR Siadenovirus OR Testadenovirus)].
The search was conducted in Web of Science Core Collection (Science Citation Index Expanded, Book Citation Index Expanded, Emerging Sources Citation Index, Current Chemical Reactions, Index Chemicus) and CAB abstracts. The outputs were merged and duplicates removed. The search was restricted to journal articles, review papers or book chapters in English, for the period 1900–2023.
As a result, 407 references were screened for title and abstract (Figure 4), 76 being selected for full text screening, and the other 331 excluded because they did not meet the inclusion criteria. Out of the 76 selected for full text screening, 57 were selected for data extraction. The other 19 were discarded because they were not in English, no full text was available or they did not meet the eligibility criteria.
Figure 4.
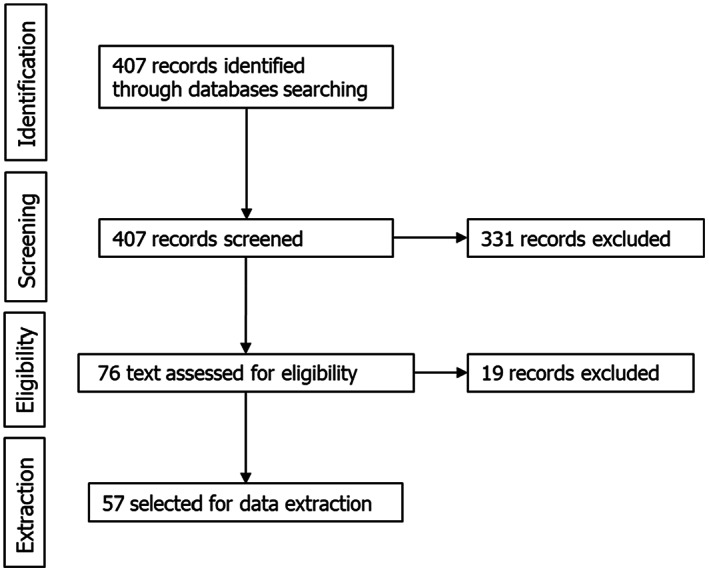
Flow diagram of the literature review for heat inactivation parameters of Parvoviridae, Circoviridae, Adenoviridae and Anelloviridae
For data extraction, the papers were screened for evidence of thermal inactivation (D‐values, z‐values, log10 reductions or any other measurement) following heat treatment with defined temperature/time combinations. The information retrieved through literature search was complemented with thermal inactivation data available for the same viral families in previous EFSA Opinions (EFSA BIOHAZ Panel, 2021). The data from the different papers were used to extract D‐values at different temperatures or, when this was not possible due to data limitations (e.g. less than three available measures), to estimate time to 1 log10 reduction based on the reduction achieved in a single point experiment. The following restrictions were applied to the data collected:
considering the technical parameters of methods 2 to 5 and five scenarios of method 7 (see Section 1.3.1) and the differences in thermal inactivation of viruses under dry and moist heat conditions (Bräuninger et al., 2000; Sauerbrei and Wutzler, 2009), only studies dealing with moist heat inactivation were included;
considering the technical parameters of methods 2 to 5 and five scenarios of method 7, only studies dealing with temperatures equal or above 50°C were considered;
taking into account the evidence on higher thermal susceptibility of Parvovirus B19 compared to other animal parvoviruses (Blumel et al., 2002; Yunoki et al., 2003), data associated to this viral species were not included.
Papers that did not provide thermal inactivation as quantitative values (e.g. reporting of ‘total inactivation’) or in which heat treatment was combined with other inactivating treatments (e.g. chemical disinfection) were also excluded. Relevant data extracted from the shortlisted references are presented in Section 3.2.4.
2.3. ‘Level of inactivation’ achieved for methods 2–5 and 7 (AQ4)
2.3.1. Calculation of the level of inactivation (log10 reduction)
Specific holding times at fixed temperatures are specified for methods 2 to 5 in Chapter III, Annex IV of Commission Regulation (EU) No 142/2011 are displayed in Table 1, and for the selected scenarios of method 7 in Table 2. Estimates of the accumulated lethality of the heat treatments were calculated for the target microorganisms: C. perfringens spores, Salmonella Senftenberg, Enterococcus faecalis and the most thermal‐resistant virus family, Parvoviridae. The level of inactivation (L) (log10 reduction) of the heat regimes was computed by Equation (1), derived from the Bigelow model (Bigelow, 1921)
| (1) |
where ttreat 1/ttreat 2/ttreat n and Ttreat 1/Ttreat 2/Ttreat n are the corresponding holding times (min) and temperatures (°C) of the n heat treatments (see Tables 1 and 2); Dref (min) is the reference D value (e.g. time to reach one log10 reduction) at the reference temperature Tref (°C); and z (°C) is the z value defined as the temperature increment needed for a 10‐fold decrease in D.
Table 1.
Time–temperature profiles corresponding to methods 2–5 in consecutive and coincidental modes providing the best‐ and worst‐case scenarios, respectively, in terms of microbial log10 reduction
| Method 2 | Method 3 | Method 4 | Method 5 | |
|---|---|---|---|---|
| Consecutive (best‐case scenario) | ||||
| Step 1 | 100°C × 125 min | 100°C × 95 min | 100°C × 16 min | 80°C × 120 min |
| Step 2 | 110°C × 120 min | 110°C × 55 min | 110°C × 13 min | 100°C × 60 min |
| Step 3 | 120°C × 50 min | 120°C × 13 min | 120°C × 8 min | – |
| Step 4 | – | – | 130°C × 3 min | – |
| Coincidental (worst‐case scenario) | ||||
| Step 1 | 100°C × 5 min | 100°C × 40 min | 100°C × 3 min | 80°C × 60 min |
| Step 2 | 110°C × 70 min | 110°C × 42 min | 110°C × 5 min | 100°C × 60 min |
| Step 3 | 120°C × 50 min | 120°C × 13 min | 120°C × 5 min | – |
| Step 4 | – | – | 130°C × 3 min | – |
Table 2.
Method 7 scenarios selected for the assessment
| Batch (B) or Continuous (C) | T (°C) | Time (min) | |
|---|---|---|---|
| Scenario 1 | C | 80 | 14 |
| Scenario 2 | C | 95 | 90 |
| Scenario 3 | B | 115 | 56 |
| Scenario 4 | C | 125 | 10 |
| Scenario 5 | B | 133 | 5 |
Taking into consideration that the steps of the heat regimes can be consecutive or coincidental in the four methods 2 to 5 (Section 3.3.1), L was estimated in the two modes: using non‐overlapping time–temperature profiles to represent the best‐case scenarios and using overlapping time–temperature profiles that comply with the heat treatment requisites but would provide the lowest log10 reduction (worst‐case scenarios) (Table 1).
In addition, five scenarios of individual time/temperature profiles corresponding to applications of method 7 were assessed (Table 2). These five time–temperature scenarios were selected based on the process parameters approved in different MS, as provided by EFPRA. As explained in the model assumptions, the effect of particle size was not taken into account in the models.
The level of inactivation in terms of log10 reductions attained was determined by performing simulations on Equation (1), using distributions for log10Dref () and z (), representing uncertainty about the heat resistance parameters of the target microorganism. The parameters and are the mean and standard deviation of the logarithm base 10 of Dref, respectively, whereas and are the mean and standard deviation of the z value, respectively. Such parameters were estimated by fitting the Bigelow model (Bigelow, 1921) of the form shown in Equation (2),
| (2) |
to literature data collected for each selected pathogen (see below the source of data and its selection). The subscripts j(i) denote the ith growth medium taken from the jth study; and εj(i) are the normally distributed residuals. Random effects with the growth medium as clustering variable were placed only in the intercept, therefore, affecting log10Dref and not z. Since extrapolation out of the temperature range available in each data set was not applied, the maximum temperature of each data set (i.e. target microorganism × inactivation method) was established as the reference temperature Tref. By doing this, the level of inactivation was estimated without recourse to extrapolation (i.e. beyond the maximum temperature available). Since distributions about log10Dref and z were built by extracting the fitted estimates and associated standard errors, these distributions represent the uncertainty about the true heat resistance parameters of the target microorganisms. Simulations were run considering the correlation between log10 Dref and z.
For each of the data sets, Bigelow models were fitted using the non‐linear mixed effects as in the nlme library of the R software (R Core Team, 2021). Monte Carlo simulations with 500,000 iterations of Equation (1) were performed using the add‐in tool for Microsoft Excel @RISK 7.6 (© 2018 Palisade Corporation).
2.3.2. Model assumptions
The model applied to assess the level of inactivation achieved by methods 2 to 5 and method 7 as stated in Regulation (EU) No 142/2011 did not take into consideration the differences among methods in terms of different maximum particle sizes, real temperatures profiles, differences in physicochemical properties of the substrates or impact of come‐up times. It was assumed that the core temperatures of the profiles for each method (either coincidental or consecutive ones) were reached at the coldest point of the pig ABP material being processed. The impact of the come‐up times (times of temperature increase or decrease before reaching the target temperature of each of the steps) on the level of inactivation was not considered, as it would mean an increased level of inactivation that cannot be quantified for each case.
It was also assumed that the survivor curves of the different reference microorganisms presented a linear shape. No shoulder or tailing effects were considered, as the D and z values were obtained from the original studies identified in the extensive literature search for each microorganism and in a previous scientific opinion (EFSA BIOHAZ Panel, 2021).
It was also assumed that the equipment used could reach the conditions specified in the legislation for all the methods. So, the evaluation performed is reliable as long as the distribution of the temperature in the equipment and the heat transfer/penetration of the matrix are taken into consideration and the T/t profile considered corresponds to the cold spot, to comply with the requirements of the Regulation (EU) No 142/2011 (i.e. minimum temperature in all material in the unit or in the reactor).
The models have been truncated so that no predictions could be obtained at temperatures higher than the maximum one for which experimental data are available. This led to an underestimation of the level of inactivation reached by some of the methods, and potentially to an underestimation of the certainty that the target log10 reductions are achieved. Equally, previous assumptions led to the underestimation (not considering the come up) or both under/overestimation (non‐linear behaviour) of the level of inactivation. All these assumptions were taken into account during the EKE.
2.3.3. Model data
Salmonella Senftenberg
Data on thermal inactivation of S. Senftenberg were obtained from the review made by Doyle and Mazzotta (2000), as in a previous EFSA scientific opinion (EFSA BIOHAZ Panel, 2021), including inactivation data for whole eggs, egg yolk, egg whites, raw milk, ground beef, nutrient broth and chocolate. The log10Dref and z distributions to be used in the determination of the level of inactivation (L) were obtained from this data set for methods 2, 3, 4 and scenarios of method 7. However, since the thermal treatment of method 5 is applied to defatted proteinaceous material, the log10 Dref and z distributions were in that case modelled using only the data for ground beef, egg whites, egg yolk, whole eggs and nutrient broth.
The parameters for log10Dref and z for use in the simulations of level of inactivation by methods 2–4, 7 and method 5 were obtained by adjusting Equation (2) to each of the two data subsets. Level of inactivation distributions were then obtained as described above. Distributions of the level of inactivation were then obtained through simulation solving the Equation (1) for each of the methods (2–5) and the 5 scenarios of method 7 for the temperature profiles shown in Tables 1 and 2.
Enterococcus faecalis
Data on thermal inactivation of E. faecalis were obtained from Ugwuanyi et al. (1999) (digestion waste), Sörqvist (2003) (mixed liquid medium), Aguirre et al. (2009) (whole milk) and Saucier and Plamondon (2011) (ground beef), as in a previous EFSA scientific opinion (EFSA BIOHAZ Panel, 2021). For E. faecalis, all available thermal inactivation data were used to adjust Equation (2) in order to determine the means and standard errors to model the uncertainty distributions about log10Dref and z. Distributions of the level of inactivation were then obtained as described above.
Spores of Clostridium perfringens
Data on D values of C. perfringens at different temperatures were obtained from Andersen et al. (2004) (Duncan and Strong medium), Brooks (2013) (fruit juice), Byrne et al. (2006) (pork luncheon roll), Craven (1990) (sodium phosphate buffer), Evelyn and Silva (2015) (beef slurry), Li et al. (2018) (laboratory medium), Ma et al. (2012) (DS medium), Orsburn et al. (2008) (DS medium), Paredes‐Sabja et al. (2008) (DS medium), Raju and Sarker (2005) (DS medium), Sarker et al. (2000) (DS medium) and Soni et al. (2022) (beef gravy). Equation (2) was fitted to these data in order to determine the means and standard errors to model the uncertainty distributions about log10Dref and z, to be used in the simulations for methods 2–4 and the 5 scenarios of method 7. For the thermal treatment of method 5, which is applied to defatted proteinaceous material, the log10Dref and z distributions were modelled using the data from pork luncheon roll, beef slurry and beef gravy. Distributions of the level of inactivation were then obtained as described above.
Parvoviruses
Data on thermal inactivation of parvoviruses were obtained from the outputs of the extensive literature search undertaken for the four families of virus selected by the hazard identification. The data from the different papers were used to extract D‐values at different temperatures (see Table B.1 of Appendix B). Available thermal inactivation data were used to adjust Equation (2) in order to determine the means and standard errors to model the uncertainty distributions about log10Dref and z. The same simulation procedure as described above was employed to obtain distributions of the level of inactivation for the heat treatment methods.
Table B.1.
Summary of the data extracted from the relevant papers on thermal inactivation parameters of members of the family Parvoviridae
| Virus | Temperature (°C) | D‐value or point estimate of time to 1 log reduction (min) | Matrix | Experimental conditions | Reference |
|---|---|---|---|---|---|
| Bovine parvovirus (BPV) | 60 | 433,0 (a) | Serum albumin |
Suspension medium: plasma. Haden strain, titre: 108 infectious units/mL. Albumin solution. |
Brauniger et al. (2000) |
| 60 | 257,8 (a) | Distilled water |
Suspension medium: AquaDest. Haden strain, titre: 108 infectious units/mL. Albumin solution. |
||
| 55 | 133,3; 576,0 (b) | Viruses on filters within solid carriers | Thermophilic fermentation. Virus (titre: 5.8 log TCID50/mL) adsorbed on filters placed within minced meat. | Paluszak et al. (2010) | |
| 55 | 150,0; 640,0 (b) | Viruses on filters within solid carriers |
Thermophilic fermentation. Virus (titre: 5.8 log TCID50/mL) adsorbed on filters placed within small meat carriers. |
||
| 55 | 600,0; 720,0 (b) | Viruses on filters within solid carriers |
Thermophilic fermentation. Virus (titre: 5.8 log TCID50/mL) adsorbed on filters placed within large meat carriers. |
||
| 55 | 240,0; 720,0 (b) | Viruses on filters within solid carriers |
Thermophilic fermentation. Virus (titre: 5.8 log TCID50/mL) adsorbed on filters placed within bone carriers. |
||
| 55 | 100,0; 443,1 (b) | Culture medium |
Thermophilic fermentation. Viral suspension (titre: 5.8 log TCID50/mL) in tubes. |
||
| 56,4 | 7236,2 (a) | Viruses on filters | Anaerobic thermophilic digestion. Virus (Haden strain) adsorbed on filters (106 PFU/filter). | Spillman et al. (1987) | |
| 60,6 | 3171,8 (a) | Viruses on filters | Aerobic thermophilic digestion. Pressure 15 kPa. Virus (Haden strain) adsorbed on filters (106 PFU/filter). Inactivation rate constant: 0,454 ± 0,018/day | ||
| 70,5 | 2087,0 (a) | Viruses on filters | Pasteurisation. Virus (Haden strain) adsorbed on filters (106 PFU/filter). Inactivation rate constant: 0,69 ± 0,018/day | ||
| Canine parvovirus (CPV) | 60 | 193,5 (b) | Blood and plasma‐derived products | Pasteurisation, albumin concentration 4% | Gröner et al. (2018) |
| 60 | 545,5 (b) | Blood and plasma‐derived products | Pasteurisation. Stabilised intermediates in plasma manufacturing processes (standard conditions). | ||
| 60 | 400,0b | Blood and plasma‐derived products | Pasteurisation. Stabilised intermediates in plasma manufacturing processes (stabiliser concentration increased to 110%). | ||
| 58 | 517,2 (b) | Blood and plasma‐derived products | Pasteurisation. Stabilised intermediates in plasma manufacturing processes (temperature reduced to 58°C). | ||
| Minute virus of mice (MVM) | 90 | 0,1; 0,2 (b) | Glucose concentrate solution | HTST pasteurisation in 50% glucose solution. Viral titre: 107 TCID50/mL | Gemmell et al. (2021) |
| Porcine parvovirus (PPV) | 60 | 73,7 (a) | Blood and plasma‐derived products | Pasteurisation. NADL‐2 strain, titre: 108 TCID50/mL | Blumel et al. (2002) |
| 52 | 1006,2 (a) | Faecal suspension | Strain 893/76, titre: 107,2 TCID50/g | Elving et al. (2014) | |
| 55 | 1336,8 (a) | Saline solution | Strain 893/76, titre: 107,2 TCID50/mL | ||
| 70 | 74,2 (a) | Saline solution | Strain 893/76, titre: 107,2 TCID50/mL | ||
| 60 | 24,7 (a) | Blood and plasma‐derived products | Pasteurisation. Viral titre: 4.88 log TCID50/mLl | Huangfu et al. (2017) |
D value.
Point estimates of time to 1 log reduction.
2.3.4. Use of inactivation parameters of spores of Clostridium botulinum and interpretation of extrapolation
Considering that the model could underestimate for some methods the level of inactivation achieved for spores of C. perfringens, as no experimental data were retrieved for this pathogen at temperatures above 105°C, estimations were also made on the level of inactivation that the methods would produce for spores of C. botulinum, considering that spores of C. botulinum are more heat resistant than spores of C. perfringens or any other food‐borne pathogenic spore‐forming bacteria (Rosnes et al., 2012).
In previous EFSA ABP scientific opinions, it was agreed to accept only scientific evidence demonstrating a sufficient level of inactivation and to disregard results of extrapolation at higher temperatures than the experimental ones. This choice was made based on evidence indicating that extrapolation beyond the experimental range can lead to serious mistakes (Masana and Baranyi, 2000; Peleg, 2021). Furthermore, the risk of extrapolation beyond the experimental limits (EFSA BIOHAZ Panel, 2022) and the difficulties to develop accurate models when experimental bias takes place (Garre et al., 2023) have also been reported recently. As a result, extrapolation of thermal inactivation beyond the conditions (in our case, temperatures) of available experimental data was not considered. Therefore, for some of the methods under assessment, the reductions that can be reached at the actual treatment temperatures of the heat regime may be underestimated.
It is accepted though, that, if the process takes place at a higher temperature than the one published in the scientific literature, the reduction/inactivation achieved would be, at least equal to that demonstrated at the lower temperature.
In previous EFSA assessments of alternative ABP processing methods applied to Category 3 ABP, if the hazard identification considers spore‐forming bacteria as relevant biological hazards, the required level of inactivation will be a 5 log10 reduction of spores from these bacteria, with the exception of spores of C. botulinum for which a 12 log10 reduction would be required to comply with a treatment equivalent to 3 min at 121°C (F0 = 3 min) 5 (Section 3.1.1). At this time/temperature conditions a 5 log10 reduction of other less heat resistant spore forming bacteria, such as C. perfringens, would be also met.
There are heat inactivation data available for C. botulinum in a wide range of temperatures and substrates, including temperatures up to 140°C (Diao et al., 2014). Therefore, the accumulated lethality expressed in minutes at Tref 121.1°C (0.25 min) of can be estimated at a wider temperature range compared to that of C. perfringens, as the z value is considered to be 10°C (Lund and Peck, 2001).
C. botulinum could be then considered as a surrogate for the assessment of the level of inactivation achieved for other relevant spore‐forming pathogens in Category 3 material, although it needs to be evaluated case by case.
2.4. AQ5: What is the certainty that the ‘level of inactivation’ achieved by methods 2, 3, 4, 5 and 7, as in AQ4, is sufficient to reach the standards for Category 3 ABP of porcine origin?
An expert knowledge elicitation (EKE) was performed to answer AQ5, based on the collected evidence and indicated uncertainties. The EKE question was specified as follows:
What is the probability that a 5 log10 reduction of spores of C. perfringens is achieved, in more than 99% of cases, by application of each of the relevant processes (methods 2, 3, 4, 5 in coincidental mode and five t/T combinations of method 7) (see Table 3), assuming that the processes are performed as prescribed and that the indicated process conditions are achieved?
Table 3.
t/T combinations assessed in the EKE exercise
| Process | t/T combinations |
|---|---|
| Method 2 – coincidental | 100°C × 5′ – 110°C × 70′ – 120°C × 50′ |
| Method 3 – coincidental | 100°C × 40′ – 110°C × 42′– 120°C × 13′ |
| Method 4 – coincidental | 100°C × 3′ – 110°C × 5′ – 120°C × 5′ – 130°C × 3′ |
| Method 5 – coincidental | 80°C × 60′ – 100°C × 60′ |
| Method 7 – scenario 1 | 80°C × 14′ |
| Method 7 – scenario 2 | 95°C × 90′ |
| Method 7 – scenario 3 | 115°C × 56′ |
| Method 7 – scenario 4 | 125°C × 10′ |
| Method 7 – scenario 5 | 133°C × 5′ |
The EKE addressed the spores of C. perfringens only because it was shown it is the most thermal resistant among the four selected indicators (Section 3.3). The processes assessed for methods 2–5 were in coincidental mode, which represent the worst‐case scenario. The processes assessed are displayed in Table 3.
In the EKE question, the phrase ‘in more than 99% of cases’ refers to the potential variation in the performance of the relevant process/es. As the process/es is/are well defined, this variation was considered to be small. The ‘probability’ refers to the certainty that the log10 reduction is achieved if this well‐defined process is performed, and not to the certainty that the conditions of the methods are applied.
The EKE consisted of two steps: individual judgements and consensus judgements.
In Step 1, the experts provided individual judgements for each of the nine processes, taking into account the version of the draft opinion at the beginning of the process with the evidence on thermal inactivation of the C. perfringens spores (including the data and modelling results), the description of the processes, the integration of the evidence and the uncertainty table (Table 9), as well as the personal expertise and assessment of the uncertainties involved.
Table 9.
Sources of uncertainty associated with the AQs and their possible impact on the conclusions
| Source of uncertainty | Cause of the uncertainty | Impact of the uncertainty on the conclusions (e.g. over/underestimation) |
|---|---|---|
| Identification of viral hazards | All viral hazards that may occur in the raw materials for production of pig PAP may not have been identified through the literature search. It is also possible that the occurrence of virus, in general, or certain families, in particular, have never been investigated in materials used for pig PAP production. | The lack of identification of a relevant virus is expected to impact the conclusions only if it would be a heat‐resistant virus (e.g. non‐enveloped DNA viruses). Parvoviruses are included in the assessment and are considered among the most heat‐resistant viruses in nature, and it is unlikely that other more heat‐resistant viruses, not considered in the opinion, could occur. |
| Technical parameters for methods 2 to 5 | The Commission Regulation (EU) 142/2011 specifies that the core temperatures listed under each method must be achieve. Either in batch or continuous mode, the temperature in different sections of the equipment must be higher in order to ensure the minimum temperatures at the core. | As estimates were made considering core temperatures being achieved, the level of inactivation achieved by processing methods 2 to 5 may be in some cases underestimated outside the core. |
| Technical parameters for method 7 | Method 7 requires compliance with microbiological criteria ensuring the non detection of Salmonella and Clostridium perfringens, and certain levels of Enterobacteriaceae, but processes approved in the EU since 2004 for method 7 have very different parameters in terms of temperature and time combinations. | Depending on the method applied, the inactivation of the indicator microorganisms in the raw material could be higher or lower than that calculated for the five scenarios included in the present opinion. |
| Data related. Identification of studies on the inactivation of the hazards | Relevant records for extraction of data on thermal inactivation of selected indicators were identified through literature searches. There is the possibility that some relevant studies were not identified or considered for data extraction. | Considering the randomness of the non‐inclusion of potentially relevant studies, this source of uncertainty could lead to either an over or underestimation of the inactivation of the biological hazards. |
| Data related. Type of matrices used for the evaluation of the inactivation of the hazards | The data extracted on the thermal inactivation of hazards were sourced from experimental studies using different matrices, in most cases of a different nature to the materials used for pig PAP production. The different composition in terms of dry matter (total solid contents, aw), fat content, etc. determines the capacity of hazards to survive under different conditions of time/temperature. | The ability of methods 2 to 5 and method 7 to achieve the targeted reductions in the materials used to produce pig PAP may be higher or lower than estimated. Most inactivation data used were derived from studies using liquid media or foods. As microbial inactivation by heat is lower in systems with lower aw, estimations from studies on liquid acidic media or on foods with high aw, this could result in an overestimation of the inactivation achieved by methods 2 to 5 and method 7. |
| Data related. Strain of the hazard and enumeration method used to assess the level of inactivation of the hazards | The data extracted on the thermal inactivation of hazards were sourced from experimental studies using specific strains/isolates of the relevant hazards and different analytical methods, which, for viral hazards, are not standardised. It is uncertain whether those strains are representative of the behaviour of the whole species. | This source of uncertainty could lead to either over or underestimate the inactivation of the biological hazards. |
| Data related. Temperature measurement used for the inactivation of the hazards | In the studies, the temperature was measured in the substrate and resembled isothermal conditions. The accuracy of the temperature measurement may affect the thermal resistance estimation. | This source of uncertainty could lead to either an over or underestimation of the inactivation of the biological hazards. |
| Data related. Heating temperatures used for the inactivation of the hazards | The data retrieved on thermal inactivation of hazards contained information on certain heating temperatures, that in some cases were far from the temperatures under assessment. Extrapolations at temperatures above those for which experimental data was available were not performed. | No extrapolation at temperatures above those for which experimental data is available may lead to an underestimation of the level of inactivation achieved by the methods, especially for those methods that work at higher regime temperatures. |
| Model related. D‐value estimation | Estimation of D values from primary research studies. D values were extracted, where available, from tables and text as in the published peer‐reviewed literature without checking the primary data. For viruses, D values from data with low linearity and point estimates of time to 1 log10 reduction at different temperatures were also included. | Underestimation or overestimation of D‐values could lead to overestimation or underestimation of log10 reductions. Uncertainty in D values introduces bidirectional uncertainty in the estimation of Dref and z. |
| Model assumptions | The model does not consider come up times. i.e. the additional inactivation due to heating and cooling times to the time/temperature combinations required for each process under assessment. | The model underestimates the level of inactivation achieved by each method, the underestimation being greater in the method/s with higher temperatures. |
| Use of Clostridium botulinum as surrogate for Clostridium perfringens | Use of thermal resistance data of C. botulinum spores, obtained in different substrates, conditions and methodologies, assuming that they will be, at least, equivalent to the thermal inactivation of C. perfringens spores in the conditions of the method evaluated. | Using D values at temperatures below those of some of the assessed methods (e.g. method 4 and method 7 scenarios 4 and 5) could lead to underestimation of the level of inactivation achieved. |
| Use of comparative analysis with Clostridium botulinum in the EKE | In the EKE both the model results and the results of the accumulated lethality in minutes at Tref 121°C of methods 2 to 5 and the scenarios of method 7, at temperature other than the reference one for C. botulinum (121.1°C), were used. | For some of the methods there was a discrepancy between the outputs of the model and of the EKE. The individual judgements may have over or underestimated the level of inactivation of the methods. |
In Step 2, during an open session, the experts were asked to consider what a rational impartial observer (RIO) would judge, having considered the evidence, uncertainties, the individual judgements and having heard the discussion maintained by the experts. The objective was to reach consensus on the probability ranges that were considered to best represent the uncertainty on whether the 5 log10 reduction of spores of C. perfringens is achieved, in more than 99% of cases, by application of each of the relevant processes. Detailed information on the EKE can be found in section 3.4 and Appendix D.
3. Assessment
3.1. What relevant pathogens can be used as indicators to assess the efficacy of standard processing methods for Category 3 ABP of porcine origin? (AQ1)
To quantify the inactivation level of each relevant pathogen (bacteria, parasites and viruses) in raw materials submitted to methods 2 to 5 and 7 is very complex because the behaviour of microorganisms throughout processing or transformation methods is difficult to elucidate for every single species and/or strain. Moreover, pathogens are irregularly distributed in the raw materials and usually occur in low prevalence and concentration. Therefore, indicator microorganisms have been used. Indicator microorganisms typically represent the most resistant pathogens within specific categories. The effect of processing or transformation methods can therefore be assessed, as if these most resistant indicator microorganisms are inactivated, then more sensitive biological hazards can also be assumed to be inactivated.
3.1.1. Criteria for selection of relevant bacterial pathogens
3.1.1.1. Previous EFSA standards applied for Category 3 animal by‐products
The EFSA scientific opinion on the evaluation of a multi‐step catalytic co‐processing hydro‐treatment for the production of renewable fuels using Category 3 animal fat and used cooking oils (EFSA BIOHAZ Panel, 2022) defined the following standards to be applied for Category 3 material:
In order to be considered at least equivalent to the processing methods approved in the legislation, the alternative methods for Category 3 ABP should be capable of reducing the concentration of the relevant pathogenic bacteria by at least 5 log 10 and the infectious titre of the relevant viruses by at least 3 log 10 (EFSA BIOHAZ Panel, 2005a). For chemical treatments, a reduction of viable stages of resistant parasites such as eggs of Ascaris sp. by at least 99.9% (3 log 10 ) shall be required. The determination of the relevant pathogenic bacteria and viruses should be defined by the hazard identification, specific for the material to be treated. If the hazard identification considers spore‐forming pathogenic bacteria to be relevant, the required level of inactivation will also be a 5 log 10 reduction of spores from these bacteria, with the exception of spores of C. botulinum for which a 12 log 10 reduction would be required, as for processing canned petfood. This is the expected reduction in C. botulinum spores after applying 121.1°C for 3 min, the minimum standard of a heat treatment for canned petfood. Given their well‐described high level of resistance to thermal and chemical treatments, applicants may choose to directly use spores of pathogenic bacteria as primary indicators without carrying out a full hazard identification exercise.
If needed/appropriate, for both spore‐forming and non‐spore‐forming bacteria and viruses, adequately justified alternative non‐pathogenic indicator or surrogate organisms with at least the same level of resistance may be used, demonstrating an equivalent level of reduction in the substrate of interest.
3.1.1.2. Legislation on alternative methods for composting and biogas
Commission Regulation (EU) No 142/2011, in Section 2.1, Chapter III, Annex V describes the alternative transformation parameters for biogas and composting plants.
The validation of the intended process referred to in point (c) must demonstrate that the process achieves the following overall risk reduction:
- for thermal and chemical processes by:
-
–a reduction of 5 log 10 of Enterococcus faecalis or Salmonella Senftenberg (775 W, H 2 S negative),
-
–reduction of infectivity titre of thermoresistant viruses such as parvovirus by at least 3 log 10 , whenever they are identified as a relevant hazard; and
-
–
- as regards chemical processes also by:
-
–a reduction of resistant parasites such as eggs of Ascaris sp. by at least 99.9% (3 log 10 ) of viable stages.
-
–
3.1.1.3. Indicators for method 7
The three microorganisms for which microbiological standards are listed in legislation for processing method 7, as described in point G.1(i) of Chapter III, Annex IV of Commission Regulation (EU) 142/2011, are: Salmonella, C. perfringens and Enterobacteriaceae.
Conclusion
Upon consideration of the three listed criteria, three bacteria were selected to be used as indicators in this assessment: S. Senftenberg, E. faecalis and spores of C. perfringens.
3.1.2. Description of the selected relevant bacterial pathogens
Salmonella Senftenberg
Salmonellae are Gram‐negative non‐spore‐forming motile rod bacteria. They are widespread in nature and found in food, soil, water, manure (Winfield and Groisman, 2003) and biological waste streams (Burtscher and Wuertz, 2003). The main reservoir of non‐typhoidal Salmonella are animals, but they are well adapted to their surroundings and cycle between environmental matrices and living hosts. Certain serovars or strains of Salmonella enterica are noted for their high resistance to thermal treatments, relative to other Salmonella spp. or Gram‐negative bacteria, the most prominent being S. Senftenberg, particularly the strain 775 W (Ng et al., 1969). In different model systems, this strain has shown D‐values (times needed to reduce the bacterial population at a given temperature by 1 log10 unit) around 10‐fold to 20‐fold higher than those of other serovars, such as Salmonella Typhimurium or Salmonella Enteritidis (Doyle and Mazzotta, 2000). S. Senftenberg is often used as an indicator organism to validate thermal treatments (Ng et al., 1969). The implication is that if a particular thermal process achieves a sufficient level of reduction for S. Senftenberg 775 W, it will also be effective against all Salmonellae and other Gram‐negative non‐spore‐forming bacteria (Doyle and Mazzotta, 2000).
Enterococcus faecalis
E. faecalis is a member of the genus Enterococcus and is a Gram‐positive non‐spore‐forming bacterium. It is described as an opportunistic pathogen, which particularly affects immunocompromised populations. E. faecalis is found in the gut of healthy humans but only reported in some warm‐blooded animals, including dogs, and chickens (Pourcher et al., 1991; Wheeler et al., 2002). E. faecalis is identified as a heat‐resistant microorganism, resulting in its successful application in process validation (Watcharasukarn et al., 2009). Indeed, E. faecalis often serves also as an indicator microorganism to characterise the performance of hygienisation processes (Sahlström, 2003). E. faecalis is the indicator organism that is mentioned in point 1 of Section 2 of Chapter III of Annex V of Commission Regulation (EU) No 142/2011. It serves as indicator microorganism for both Gram‐positive and Gram‐negative non‐spore‐forming bacteria, given the higher thermal tolerance that Gram‐positive cocci generally show as compared with that of other non‐spore‐forming bacterial species.
Spores of Clostridium perfringens
C. perfringens is a Gram‐positive square‐ended anaerobic (microaerophilic) bacillus classified in Group III of the Family Bacillaceae (EFSA, 2005a,b). This non‐motile member of the clostridia forms oval, central spores rarely seen in culture unless grown in specially formulated media, although the spores are produced readily in the intestine (EFSA, 2005a,b). C. perfringens is ubiquitous and widely distributed in soil, dust, vegetation and raw foods. It is part of the normal flora of the intestinal tract of humans and animals. C. perfringens was first recognised as being responsible for food poisoning in the 1940s and currently is a leading cause of food‐borne illnesses. The symptoms consist of diarrhoea and abdominal cramps and appear 8–24 h following ingestion of large numbers of vegetative cells in temperature‐abused protein‐based foods. Cells sporulate in the small intestine, producing an enterotoxin. C. perfringens strains are classified into five toxicological types (A–E) based on the four major toxins produced (alpha (α), beta (β), epsilon (ε) and iota (ι)). Most of the strains produce α‐toxin (lecithinase, phospholipase C). Only C. perfringens belonging to types A and C are able to cause human gastroenteritis. C. perfringens is one of the microbiological standards listed in the EU Regulation 142/2011 for method 7 and should be absent in 1 g of product tested directly after the treatment.
3.1.3. Criteria for selection of relevant viral pathogens
Three criteria have been applied to select the relevant viral pathogens:
To be included in the WOAH list of swine and multiple species diseases
OR
-
2
to be included in the AHAW Scientific Opinion on the assessment of control measures of the Category A diseases of the Animal Health Law (AHL) (EFSA AHAW Panel, 2022)
OR
-
3
to be identified in an ELS on virus presence in pigs AND (to be present in the EU OR pose significant risk of introduction into the EU AND being pathogens to humans or animals)
The following viral pathogens were included: foot and mouth disease virus, African swine fever virus, classical swine fever virus, because they are named in the AHL as category A listed diseases and considered in the Scientific Opinion on the assessment of control measures of the Category A diseases of the Animal Health Law (EFSA, 2022); porcine reproductive & respiratory syndrome virus, porcine epidemic diarrhoea virus, Aujeszky's disease virus, swine influenza virus, because they are named in the Annex II of Regulation (EU) 2016/429 or in the WOAH list of swine diseases or diseases of multiple species (including swine). Senecavirus, as presenting a current threat to the EU due to the presence in the region.
For viral hazard identification, occurrence of the viral pathogens in pigs at slaughtering or in pig tissues or in pig products (i.e. considered fit for human consumption) was used as a proxy for their occurrence in Category 3 ABP. An ELS was conducted considering viruses occurring in pig husbandry in the EU or in third countries; viruses not detected in the EU and with a marginal likelihood of introduction in the EU through pig by‐products (e.g. Ebola viruses) were not specifically considered in the search. Viruses were considered relevant if pathogenic to humans (e.g. through the introduction in the food chain) or to animals, including poultry, fish and pigs.
The result of the ELS and screening for the viral hazards is given in Table 4. Studies dealing with seroprevalence, experimental infections or with no detection of viral pathogens were excluded.
Table 4.
Summary of the data extraction in the literature review for viral hazard identification
| Family | Characteristics | Viral hazard | Matrix | References |
|---|---|---|---|---|
| Adenoviridae | Non‐enveloped (linear) dsDNA | Porcine adenovirus (PAdV) | Spleen | Kadoi et al. (1997) |
| Swab slaughter line (bleeding) | Jones and Muehlhauser (2017a) | |||
| Faeces, liver, muscle, Sausages | Berto et al. (2012) | |||
| Faeces, liver, packaged meat, sausages | Di Bartolo et al. (2012) | |||
| Nasal/faecal swabs pools | Hause et al. (2016a) | |||
| Anelloviridae | Non‐enveloped (circular) ssDNA | Torque Teno virus (TTSuV) | Liver, serum, lung | de Arruda Leme et al. (2013) |
| Liver, pork | Leblanc et al. (2014) | |||
| Slaughterhouse collected fetuses | Martinez‐Guino et al. (2010) | |||
| Nasal/faecal swabs pools | Hause et al. (2016a) | |||
| Torque Teno virus 1 (TTSuV1) | Bile, fresh pork liver sausages | Monini et al. (2016) | ||
| Kidney | Ghosh et al. (2018) | |||
| Liver | Da Silva et al. (2020) | |||
| Blood | Luka et al. (2016) | |||
| Lymph nodes | Huang et al. (2013) | |||
| Torque Teno virus 2 (TTSuV2) | Liver | Da Silva et al. (2020) | ||
| Arteriviridae | Enveloped ssRNA+ | Porcine reproductive and respiratory syndrome virus (PRRS) | Serum | Almeida et al. (2018) |
| Meat sample from carcass, serum | Magar and Larochelle (2004) | |||
| Tonsils | O'Sullivan et al. (2011) | |||
| Lung | Hillen et al. (2014) | |||
| Cranioventral lobe, dorsocaudal lobe, heart, kidney, liver, lymph nodes, small intestine, spleen, testis, tonsils | Ho et al. (1999) | |||
| Nasal/faecal swabs pools | Hause et al. (2016a) | |||
| Spray‐dried porcine plasma | Blázquez et al. (2022) | |||
| Asfarviridae | Enveloped (linear) dsDNA | African Swine Fever (ASF) | Serum | Gallardo et al. (2011) |
| Tissue, blood | Cho et al. (2022) | |||
| Tissue | Abworo et al. (2017) | |||
| Blood | Thomas et al. (2016) | |||
| Tissue | Owolodun et al. (2010) | |||
| Blood | Adedeji et al. (2022) | |||
| Liver, lymph nodes, spleen | Owolodun et al. (2007) | |||
| Blood | Ebwanga et al. (2022) | |||
| Blood | Luka et al. (2016) | |||
| Not reported | Kong et al. (2021) | |||
| Astroviridae | Non‐enveloped ssRNA+ | Astrovirus (AstV) | Faeces | Machnowska et al. (2014) |
| Porcine astrovirus | Faeces | Luo et al. (2011) | ||
| Nasal/faecal swabs pools | Hause et al. (2016a) | |||
| Caliciviridae | Non‐enveloped ssRNA+ | Porcine enteric calicivirus (PEC) | Faeces | Halaihel et al. (2010) |
| Retail pork | Jones and Muehlhauser (2017b) | |||
| Calicivirus | Nasal/faecal swabs pools | Hause et al. (2016a) | ||
| European swine norovirus (NOV) | Faeces | Machnowska et al. (2014) | ||
| Norovirus | Faeces | Laconi et al. (2020) | ||
| Circoviridae | Non‐enveloped (circular) ssDNA | Porcine circovirus | Nasal/faecal swabs pools | Hause et al. (2016a) |
| Porcine circovirus Type 1 (PCV1) | Serum | Quintana et al. (2001) | ||
| Not reported | Csagola et al. (2008) | |||
| Kidney, lung, lymph nodes, spleen | Hu et al. (2022) | |||
| Liver | Da Silva et al. (2020) | |||
| Porcine circovirus Type 2 (PCV2) | Tonsils | Saekhow et al. (2015) | ||
| Not reported | Csagola et al. (2008) | |||
| Lymph nodes | Ojok et al. (2013) | |||
| Tissue | Jia et al. (2022) | |||
| Lymph nodes | Laisse et al. (2018) | |||
| Kidney, lung, lymph nodes, spleen | Hu et al. (2022) | |||
| Kidney | Ghosh et al. (2018) | |||
| Faeces, kidney | Kleymann et al. (2020) | |||
| Liver | Da Silva et al. (2020) | |||
| Tonsils | O'Sullivan et al. (2011) | |||
| Lung | Hillen et al. (2014) | |||
| Lung | Yue et al. (2022) | |||
| Plasma | Blazquez et al. (2019) | |||
| Spray‐dried porcine plasma | Blázquez et al. (2022) | |||
| Porcine circovirus Type 2a (PCV2a) | Plasma | Shen et al. (2011) | ||
| Porcine circovirus Type 2b (PCV2b) | Plasma | Shen et al. (2011) | ||
| Porcine circovirus Type 3 (PCV3) | Tissue | Jia et al. (2022) | ||
| Kidney, lymph nodes | Hu et al. (2022) | |||
| Lung | Wen et al. (2018) | |||
| Lymph nodes | Liu et al. (2019) | |||
| Lung | Yue et al. (2022) | |||
| Porcine circovirus Type 4 (PCV4) | Lung | Yue et al. (2022) | ||
| Coronaviridae | Enveloped ssRNA+ | Porcine epidemic diarrhoea virus (PEDV) | Swabs from lower floor of truck after animal unloading at slaughterhouse | Boniotti et al. (2018) |
| Nasal/faecal swabs pools | Hause et al. (2016a) | |||
| Spray‐dried porcine plasma | Blázquez et al. (2022) | |||
| Porcine hemagglutinating encephalomyelitis virus | Nasal/faecal swabs pools | Hause et al. (2016a) | ||
| Porcine deltacoronavirus (PDCoV) | Nasal/faecal swabs pools | Hause et al. (2016a) | ||
| Flaviviridae | Enveloped ssRNA+ | Classical swine fever (CSF) | Tissue | Sarma and Meshram (2008) |
| Spleen | Rout and Saikumar (2016) | |||
| Tissue | Rout et al. (2015) | |||
| Tissue | Sarma et al. (2008) | |||
| Intestine, kidney, lymph nodes, spleen, tonsils | Sarma et al. (2007) | |||
| Atypical porcine pestivirus | Nasal/faecal swabs pools | Hause et al. (2016a) | ||
| Hepadnaviridae | Enveloped partially‐dsDNA | Hepatitis B virus (HBV) | Bile, liver | Vieira et al. (2014) |
| Hepeviridae | Quasi‐enveloped* (non‐enveloped) ssRNA+ | Hepatitis E virus (HEV) | Liver | Jori et al. (2016) |
| Faeces | Jones and Johns (2012) | |||
| Liver | Muller et al. (2017) | |||
| Liver | Bouquet et al. (2011) | |||
| Swab slaughter line | Lainšček et al. (2017) | |||
| Faeces | Machnowska et al. (2014) | |||
| Liver | Gutierrez‐Vergara et al. (2015) | |||
| Faeces | Lainšček et al. (2017) | |||
| Blood, kidney, liver | Geng et al. (2019) | |||
| Faeces | Di Martino et al. (2010) | |||
| Faeces | Cappai et al. (2018) | |||
| Bile | Wang et al. (2015) | |||
| Liver | Vonlanthen‐Specker et al. (2021) | |||
| Pork liver surface | Dzierzon et al. (2022) | |||
| Liver | Traore et al. (2015) | |||
| Liver | de Paula et al. (2013) | |||
| Bile, liver | Gardinali et al. (2012) | |||
| Blood | Sooryanarain et al. (2020) | |||
| Bile, faeces, liver pork, meat | Intharasongkroh et al. (2017) | |||
| Faeces liver sausages | Berto et al. (2012) | |||
| Faeces, liver, packaged meat, sausages | Di Bartolo et al. (2012) | |||
| Bile | dos Santos et al. (2011) | |||
| Bile | Mughini‐Gras et al. (2017) | |||
| Faeces, liver | Chambaro et al. (2021) | |||
| Bile | Amorim et al. (2018) | |||
| Bile, bladder, faeces, liver, lymph nodes, plasma, tonsils | Leblanc et al. (2010) | |||
| Diaphragm, faeces, liver | Chelli et al. (2021) | |||
| Not reported, serum | de Souza et al. (2012) | |||
| Blood, rectum swabs | Khounvisith et al. (2018) | |||
| Liver | Feurer et al. (2018) | |||
| Caecum content, liver, serum | Boxman et al. (2022) | |||
| Liver | Temmam et al. (2013) | |||
| Liver | Rose et al. (2011) | |||
| Liver | Motoya et al. (2019) | |||
| Bile, liver | Casas et al. (2011) | |||
| Liver | Pellerin et al. (2022) | |||
| Bile | Zhang et al. (2018) | |||
| Faeces, liver | Forero et al. (2017) | |||
| Blood, liver | Bigoraj et al. (2021) | |||
| Blood | Boxman et al. (2017) | |||
| Faeces, plasma | Leblanc et al. (2007) | |||
| Liver | Wang et al. (2023) | |||
| Liver, swab slaughter line | Milojević et al. (2019) | |||
| Pork liver surface | Li et al. (2011) | |||
| Herpesviridae | Enveloped (linear) dsDNA | Porcine cytomegalovirus (PCMV) | Lung | Tajima and Kawamura (1998) |
| Orthomyxoviridae | Enveloped segmented ssRNA‐ | Influenza A (H1N1, pandemic strain 2009) | Lung | Paladino et al. (2017) |
| Nasal swab | Perera et al. (2013) | |||
| Blood | Nokireki et al. (2013) | |||
| Nasal swabs | Chen et al. (2021) | |||
| Lung | Olaniyi et al. (2020) | |||
| Nasal swabs | Rose et al. (2013) | |||
| Nasal swabs | Takemae et al. (2017) | |||
| Nasal swabs | Baudon et al. (2018) | |||
| Serum/nasal swab | Baudon et al. (2015) | |||
| Nasal swabs/tracheal swab | Cheung et al. (2022) | |||
| H7N2 influenza virus | Lung | Kwon et al. (2011) | ||
| H1N2 influenza virus | Nasal swabs | Qiao et al. (2014) | ||
| Influenza virus | Nasal swabs | Osoro et al. (2019) | ||
| Nasal/faecal swabs pools | Hause et al. (2016a) | |||
| Lung | De Conti et al. (2021) | |||
| Tracheal swab | Meseko et al. (2018) | |||
| Tracheal swab | Amorim et al. (2013) | |||
| Nasal swabs | Ducatez et al. (2015) | |||
| Swine influenza virus (SIV) | Lung | Kwon et al. (2011) | ||
| Lung | Florez et al. (2018) | |||
| Lung | Hillen et al. (2014) | |||
| Nasal swabs | Papatsiros et al. (2015) | |||
| Nasal swabs | Oladipo et al. (2013) | |||
| Nasal swabs | Baudon et al. (2018) | |||
| Spray‐dried porcine plasma | Blázquez et al. (2022) | |||
| Paramyxoviridae | Enveloped ssRNA‐ | Porcine parainfluenza virus type 1 (PPIV‐1) | Assorted tissue, bronchoalveolar lavage, lung, nasal swabs, nasal turbinate, oral fluid, respiratory swab | Park et al. (2019) |
| Parvoviridae | Non‐enveloped (linear) ssDNA | Bocavirus | Faeces, lymph nodes, nasopharyngeal swab, serum | Lau et al. (2011) |
| Inguinal lymph node, spleen, submandibular lymph node, tonsils | Liu et al. (2014) | |||
| Liver | Da Silva et al. (2020) | |||
| Nasal/faecal swabs pools | Hause et al. (2016a) | |||
| Porcine boca‐like virus (PBo‐likeV) | Tonsils | Saekhow et al. (2015) | ||
| Hokovirus | Nasal/faecal swabs pools | Hause et al. (2016a) | ||
| Porcine parvovirus | Heart | Streck et al. (2013) | ||
| Tonsils | Streck et al. (2013) | |||
| Tonsils | Saekhow et al. (2015) | |||
| Fetuses at abattoir | Mengeling et al. (1991) | |||
| Follicular fluid | Pogranichniy et al. (2008) | |||
| Kidney | Ghosh et al. (2018) | |||
| Liver | Da Silva et al., 2020 | |||
| Nasal/faecal swabs pools | ||||
| Spray‐dried porcine plasma | Blázquez et al. (2022) | |||
| Porcine parvovirus 1 (PPV1) | Blood, lung | Thuy et al. (2021) | ||
| Porcine parvovirus 2 (PPV2) | Heart | Streck et al. (2013) | ||
| Tonsils | Streck et al. (2013) | |||
| Tonsils | Saekhow et al. (2015) | |||
| Blood | Thuy et al. (2021) | |||
| Lung | Thuy et al. (2021) | |||
| Porcine parvovirus 3 (PPV3) | Tonsils | Streck et al. (2013) | ||
| Tonsils | Saekhow et al. (2015) | |||
| Blood | Thuy et al. (2021) | |||
| Lung | Thuy et al. (2021) | |||
| Porcine parvovirus 4 (PPV4) | Tonsils | Streck et al. (2013) | ||
| Tonsils | Saekhow et al. (2015) | |||
| Lung | Thuy et al. (2021) | |||
| Porcine parvovirus 7 (PPV7) | Lung | Chen et al. (2018) | ||
| Picobirnaviridae | Non‐enveloped segmented dsRNA | Picobirnavirus | Nasal/faecal swabs pools | Hause et al. (2016a) |
| Picornaviridae | Non‐enveloped ssRNA+ | Encephalomyocarditis virus (EMCV) | Faeces | Machnowska et al. (2014) |
| Enterovirus | Nasal/faecal swabs pools | Hause et al. (2016a) | ||
| Foot‐and‐mouth‐disease virus (FMDV) | Heart | Sharmila and Sherikar (2005) | ||
| Kobuvirus | Nasal/faecal swabs pools | Hause et al. (2016a) | ||
| Parecho‐like virus | Nasal/faecal swabs pools | Hause et al. (2016a) | ||
| Pasivirus | Nasal/faecal swabs pools | Hause et al. (2016a) | ||
| Sapelovirus | Nasal/faecal swabs pools | Hause et al. (2016a) | ||
| Intestinal content | Swati et al. (2020) | |||
| Senecavirus A | Nasal/faecal swabs pools | Hause et al. (2016b) | ||
| Nasal/faecal swabs pools | Hause et al. (2016b) | |||
| Senecavirus A SVA‐GD5‐2018, SVA‐GDSZ‐2018 | Lymph nodes | Jiang et al. (2021) | ||
| Teschovirus | Nasal/faecal swabs pools | Hause et al. (2016a) | ||
| Intestinal content | John et al. (2022) | |||
| Swab slaughter line (bleeding) | Jones and Muehlhauser (2017a) | |||
| unclassified Picornavirales | Posavirus | Nasal/faecal swabs pools | Hause et al. (2016a) | |
| Reoviridae | Non‐enveloped segmented dsRNA | Porcine rotavirus (RV) | Swab slaughter line (bleeding) | Jones and Muehlhauser (2017b) |
| Rotavirus group A (GARV) | Faeces | Machnowska et al. (2014) | ||
| Rotaviruses | Faeces | Halaihel et al. (2010) | ||
| Tobaniviridae | Enveloped ssRNA+ | Torovirus | Nasal/faecal swabs pools | Hause et al. (2016a) |
‘Quasi‐enveloped’ viruses (Feng et al., 2014) are viruses historically considered as non‐enveloped due to the features displayed at excretion and in the environment that, however, circulate in the bloodstream of an infected subject in a membrane‐cloaked form. This attribute has been described, among others, in Hepatitis E virus (Takahashi et al., 2010).
Resistance to thermal and chemical processes is generally accentuated in non‐enveloped viruses, rather than in enveloped ones (McDonnell, 2020). Therefore, the identification of relevant viral hazards focused on families displaying a viral and genome structure associated with higher thermal resistance (i.e. absence of envelope, DNA genome).
The following non‐enveloped DNA viruses were therefore identified as relevant hazards:
porcine adenovirus (Adenoviridae),
Torque Teno virus (Anelloviridae),
porcine circovirus (Circoviridae),
bocavirus (Parvoviridae),
porcine parvovirus (Parvoviridae).
3.1.4. Description of the pre‐selected relevant viral pathogens
Family Adenoviridae
Porcine adenovirus (PAdV) is a species of the Mastadenovirus genus within the Adenoviridae family, with a double‐stranded DNA genome of ~ 31–34 kbp. The virion displays an icosahedral symmetry with protruding fibres and a diameter of ~ 80 nm. Five serotypes divided in three species (Porcine mastadenovirus A with serotypes 1 to 3, Porcine mastadenovirus B with serotype 4 and Porcine mastadenovirus C with serotype 5) have been described to date (Benkő et al., 2022).
PAdV is considered a low‐grade porcine pathogen, mostly associated with subclinical/mild and transitory infections and with limited impact on swine production. Clinical manifestations, where present, include gastrointestinal symptoms (diarrhoea, dehydration, etc.), particularly in association with PAdV‐A or respiratory disease, encephalitis and reproductive disorders, including abortion in sows (Berto et al., 2012; Di Bartolo et al., 2012; Benfield and Hesse, 2019). While a possible role of PAdV in coinfections with other severe porcine pathogens has been hypothesized, some serotypes of PAdV have been significantly exploited for the development of recombinant vaccines towards economically relevant swine diseases (Tuboly and Nagy, 2001). The occurrence of the virus in pigs at the slaughtering time is variable, with a detection rate close to 100% in faeces and slaughtering environments and ranging from 1% to 40% in different pig tissues/organs (Berto et al., 2012; Di Bartolo et al., 2012).
Family Anelloviridae
The family Anelloviridae includes heterogeneous single‐stranded circular DNA virus (2–3.9 kb), grouped in genera named following the letters of the Greek alphabet (Alphatorquevirus, Betatorquevirus, etc.). Anelloviruses were first described in 1997 (Nishizawa et al., 1997) and, based on genome structure and morphological similarity, were previously classified in the family Circoviridae. Two species of porcine anelloviruses have been reported, torque teno sus virus 1 (TTSuV1, with two genotypes) and torque teno sus virus 2 (TTSuV2, with three genotypes), which display low nucleotide identity (Cortey et al., 2011). TTSuV transmission is mainly faecal‐oral, though vertical transmission has also been demonstrated. The infection by TTSuV is usually subclinical in swine and their pathogenic potential is still unclear, but a role in the development and the outcome of some diseases such as post‐weaning multisystemic wasting syndrome has been hypothesized (Kekarainen and Segales, 2012). The detection rate of TTSuVs in swine populations is usually high, ranging from 17% in blood and lymph nodes to almost 100% in liver (Huang et al., 2013; de Arruda Leme, 2013; Leblanc et al., 2014).
Family Circoviridae
Porcine circoviruses (PCV) (genus Circovirus) are single‐stranded circular DNA viruses with genomes of ~ 1.7–2.0 kb and a virion of 20–25 nm in diameter. Four circovirus genotypes have been detected in pigs: porcine circovirus 1 (PCV1), considered non‐pathogenic; porcine circovirus 2 (PCV2, divided in four subtypes PCV2a, PCV2b, PCV2c, PCV2d), the predominant pathogen, responsible for the different syndromes collectively described as porcine circovirus‐associated diseases (PCVADs); porcine circovirus 3 (PCV3), described in 2015 and associated to porcine dermatitis and nephropathy syndrome (PDNS); and porcine circovirus 4 (PCV4), discovered in 2019 and also associated to porcine dermatitis and nephropathy syndrome (PDNS). The PCVADs induced by PCV2 occur frequently in weaning piglets, leading to a progressive loss of weight and of body condition, with a pathogenicity mechanism not completely understood yet. The detection of PCV in pigs at slaughtering is commonly reported, with virus detection in plasma, lymph nodes, kidneys, spleen, lung and liver (Shen et al., 2011; Hu et al., 2022; Kleyman et al., 2020; Da Silva et al., 2020).
Family Parvoviridae
Porcine parvovirus (genus Protoparvovirus) and bocavirus (genus Bocaparvovirus) are members of the family Parvoviridae, subfamily Parvovirinae, that includes the species infecting vertebrates. Parvoviridae are small (25–30 nm), resistant, non‐enveloped viruses with linear, single‐stranded DNA genomes of 4–6 kb. P. parvovirus (currently designated as ungulate protoparvovirus 1, PPV1) is the causative agent of the SMEDI syndrome (stillbirths, mummification, embryonic death and infertility), a highly impacting disease in pig husbandry. Infection of seronegative gilts or sows during pregnancy leads – depending on the stage of gestation – to the typical manifestations of the disease. In the last decade, several new porcine parvoviruses have been recognised (Cadar et al., 2013) and provisionally named Porcine parvovirus 2 to 6 (PPV2, PPV3, etc.). Although the occurrence of the disease has significantly reduced due to vaccination, porcine parvoviruses are geographically ubiquitarian, and they are frequently detected in slaughtered pigs (e.g. depending on the type of porcine parvovirus, 7–78% in tonsils, 55–60% in heart tissue, 8–68% in lungs, 34% in blood (Streck et al., 2013; Thuy et al., 2021)). Porcine bocavirus (PboV, reviewed in Aryal and Liu, 2021) was discovered in 2009 in Sweden in pigs suffering from post‐weaning multisystemic wasting syndrome (PMWS) (Blomström et al., 2010). Six groups are described (PboV1 to PboV6V7V) based on VP1 sequence. Due to the detection of PboV in co‐infection with other viruses, the pathogenesis of these viruses has not been determined yet, though its occurrence has been reported in several tissues, including lymph nodes, spleen, liver, as well as in faeces (Liu et al., 2014; Da Silva et al., 2020).
3.1.5. Criteria for selection of relevant parasitic pathogens
Two criteria have been applied to select the relevant parasitic pathogens:
To be included in the WOAH list of swine and multiple species diseases OR to be included in the AHAW Scientific Opinion on the assessment of control measures of the Category A diseases of the Animal Health Law (EFSA, 2022).
Two parasites fulfil these criteria: Taenia solium and Trichinella spp.
3.1.6. Description of the selected relevant parasitic pathogens
Trichinella spp.
Trichinella constitutes a genus of worldwide distributed parasitic nematodes (roundworms). Most of the 13 genotypes described to date are zoonotic, with all of them being able to infect humans and domestic pigs (Cristómoro‐Jorquera and Landaeta‐Aqueveque, 2022). Other domestic and wild animals such as wild boars, horses, bears, rodents or foxes can also be reservoirs. Two life cycles have been described: the domestic cycle involves pigs and rodents as hosts, the sylvatic cycle includes wild mammals, birds and reptiles. Humans are infected by consumption of raw or undercooked contaminated meat, especially game meat and pork. In Europe, pigs, horses and wild boars are the main sources of infection. The transmission between non‐human animals occurs by predation or carrion consumption. The infective larvae are located within the muscle cells and, after consumption, are liberated in the stomach to later become adult worms in the small intestine. Following mating, new larvae progenies move to the muscle. Depending on the number of infective larvae ingested, symptomatology ranges from mild (fatigue, weakness, fever, diarrhoea, muscle pains) to severe (heart or breathing problems and, rarely, death) (Franssen et al., 2021; Gabriel et al., 2023). Symptoms appear from 1 to 2 days after consumption until 2–8 weeks after treatment (European Centre for disease control (ECDC 6 )). While the incidence of trichinosis from pig consumption has decreased lately, that coming from game meat has simultaneously increased (Cristómoro‐Jorquera and Landaeta‐Aqueveque, 2022). Cooking, freezing and irradiation are the three admitted processing methods for Trichinella inactivation before human consumption (Franssen et al., 2021). The EU legislation 7 suggests freezing at −21°C for 7 days for complete inactivation of T. spiralis in pork. A heat‐inactivation model proposed cooking at 60°C for 10 min for Trichinella muscle larvae inactivation (Franssen et al., 2019, 2021). Prevention includes mandatory inspection of all slaughtered pigs and horses in the EU.
Taenia solium
T. solium is a pork tapeworm found in pigs, as the intermediate host, and humans as the sole definitive host. Pigs are infected by ingesting tapeworm eggs from faeces, which subsequently develop in the muscles, eyes and the central nervous system. Human infection occurs by eating raw or undercooked infected pork containing viable tapeworm larvae (cysticerci). In its adult form T. solium causes taeniasis, whose symptomatology is usually absent or mild including abdominal pain, nausea, diarrhoea or constipation (WHO 8 , Gabriel et al., 2023). Additionally, ingestion of tapeworm eggs (via the faecal‐oral route or contaminated food or water) may lead to infection with the larval parasite in the tissues (cysticercosis). Human cysticercosis affects muscles, skin, eyes and central nervous system. When cysts develop in the brain the disease is referred as to neurocysticercosis, able to cause seizures, convulsions, blindness and even death. T. solium is endemic in large parts of Asia, Latin America and sub‐Saharan Africa (Jansen et al., 2021). Its control includes proper hygiene, preventive chemotherapy (Haby et al., 2020), treatment of pigs and meat inspection (Carabin and Traore, 2014). For elimination, T. solium cysticerci in pork are inactivated by freezing at −24 to −5°C for 1–4 days or cooking of pork meat at 60°C for a minimum period of time (Franssen et al., 2019).
The collection of the thermal resistance parameters for parasites was excluded ‘a priori’ since it is well known that the thermal resistance of parasites is lower than that of the bacterial and viral indicators selected.
3.2. What are the thermal inactivation parameters (D and z) of the biological hazards identified in AQ1? (AQ3)
3.2.1. Spores of Clostridium perfringens
An extensive literature search (ELS) was performed to collect data on the thermal inactivation of spores of C. perfringens in different substrates. The search revealed a range of D and z parameters for this bacterium in different conditions. A total of 91 D‐values were collected in the temperature range from 85 to 105°C. The information is summarised in Table A.1 of Appendix A.
Table A.1.
Summary of the data extracted from the selected papers in the literature review for the identification of thermal inactivation parameters of spores of Clostridium perfringens expressed as D values
| Temperature | D‐value (minutes) | Matrix | Pre‐treatment | Comment | Reference |
|---|---|---|---|---|---|
| 85 | 19 | Duncan and Strong sporulation media | Spores produced at 37°C | Treatment in an immersed‐coil heating apparatus to immediately heat at 85°C; strain with a chromosomal cpe gene | Andersen et al. (2004) |
| 2.10 | |||||
| 17 | Spores produced at 42°C | ||||
| 1 | |||||
| 90 | 30.6 | Pork luncheon roll | Treatment in a temperature‐controlled water bath; Three‐strain cocktail used | Byrne et al. (2006) | |
| 95 | 9.7 | ||||
| 100 | 1.9 | ||||
| 95 | 35 | Sodium phosphate buffer | Spores produced in in DS‐MOPS pH 7.0 | Recovery on BASE + L medium; treatment in glass capillary tubes |
Craven (1990) |
| 105 | 1.9 | ||||
| 95 | 75 | Spores produced in in DS‐MOPS pH 7.5 | |||
| 105 | 5.9 | ||||
| 95 | 48 | ||||
| 105 | 0.13 | ||||
| 95 | 30 | Spores produced in in DS‐EPPS pH 7.5 | |||
| 105 | 0.19 | ||||
| 95 | 40 | ||||
| 105 | 3.8 | ||||
| 95 | 56 | Spores produced in in DS‐MOPS pH 8.0 | |||
| 105 | 0.13 | ||||
| 95 | 128 | ||||
| 105 | 20 | ||||
| 95 | 84 | Spores produced in in DS‐EPPS pH 8.0 | |||
| 105 | 6.4 | ||||
| 95 | 50 | Spores produced in in DS‐EPPS pH 8.5 | |||
| 95 | 144 | ||||
| 105 | 6.4 | ||||
| 95 | 50 | Spores produced in in DS‐EPPS pH 8.5 | Recovery on BASE medium; treatment in glass capillary tubes | ||
| 105 | 0.22 | ||||
| 95 | 32 | Spores produced in in DS‐EPPS pH 8.0 | |||
| 105 | 0.16 | ||||
| 95 | 28 | Spores produced in in DS‐MOPS pH 7.0 | |||
| 105 | 0.13 | ||||
| 105 | 2.5 | Beef slurry, 14% protein, 7% fat | Spores produced in modified Duncan‐strong (DS) sporulation medium | Treatment in thin layer pouches (1‐2 mm × 8 cm × 8 cm), submerged in oil bath. No info of actual temp in the centre is provided | Evelyn and Silva (2015) |
| 100 | 7.1 | ||||
| 95 | 21.7 | ||||
| 105 | 1.8 | Beef slurry | Spores produced in modified Duncan‐strong (DS) sporulation medium | Treatment in thin layer pouches (1‐2 mm × 8 cm × 8 cm), submerged in oil bath. | |
| 100 | 5.5 | ||||
| 95 | 15 | ||||
| 100 | 0.5 | Unclear, water or laboratory broth | In: Sarker MR, Shivers RP, Sparks SG, Juneja VK, McClane BA (2000) Comparative experiments to examine the effects of heating on vegetative cells and spores of Clostridium perfringens isolates carrying plasmid versus chromosomal enterotoxin genes. Appl Environ Microbiol 66: 3234–3240 | Not indicated how it was done, values for several strains. | Li and McClane (2008) |
| 10 | |||||
| 59.1 | |||||
| 8.7 | |||||
| 44.7 | |||||
| 16.4 | |||||
| 9.3 | |||||
| 38 | |||||
| 50 | |||||
| 0.7 | |||||
| 100 | 13.5 | MDS medium | Heat treatment very vague, immersion in boiling water of 10 mL tubes | ||
| 16.6 | |||||
| 40.3 | |||||
| 20.8 | |||||
| 20.5 | |||||
| 33.4 | |||||
| 7 | |||||
| 100 | 16 | Distilled water | Spores produced in Duncan and Strong (DS) sporulation medium | Heat treatment in tubes, exposure not specified to 100°C, poor description of the methodology | Novak and Yuan (2003) |
| 90 | 120.6 | Not specified, laboratory conditions | Spores produced in Duncan and Strong (DS) sporulation medium | Method not explained, in: Ando, Y., T. Tsuzuki, H. Sunagawa, and S. Oka. 1985. Heat resistance, spore germination, and enterotoxigenicity of Clostridium perfringens. Microbiol. Immunol. 29:317–326 | Osburn et al. (2008) |
| 45.6 | |||||
| 21.4 | |||||
| 19.9 | |||||
| 19 | |||||
| 12.5 | |||||
| 10.1 | |||||
| 6.9 | |||||
| 5.5 | |||||
| 100 | 49.1 | DS medium | Spores produced in Duncan and Strong (DS) sporulation medium | Heat treatment at 100°C, not clearly explained, submerged | Paredes‐Sabja et al. (2008) |
| 19.2 | |||||
| 45.2 | |||||
| 28.8 | |||||
| 52.2 | |||||
| 100 | 62 | As Sarker et al. (2000) | Same as Sarker et al. (2000) | Heat treatment at 100°C, not clearly explained, submerged | Raju and Sarker (2005) |
| 61 | |||||
| 0.5 | |||||
| 100 | 124 | DS medium culture | Spores in Duncan and Strong sporulation | Flask technique or similar, temp fixed at 90 or 100°C, then spore suspension added, mixed and sampled at time intervals | Sarker et al. (2000) |
| 67 | |||||
| 32 | |||||
| 30 | |||||
| 45 | |||||
| 60 | |||||
| 0.5 | |||||
| 1.9 | |||||
| 1.6 | |||||
| 0.9 | |||||
| 1.3 | |||||
| 0.5 | |||||
| 104 | 2.9 | Commercial beef gravy | Not available | Data from Bradshaw JG, Peeler JT and Twedt RM, 1977. Thermal inactivation of ileal loop‐reactive Clostridium perfringens type A strains in phosphate buffer and beef gravy. Applied Environmental Microbiology, 34, 280–284. Borosilicate glass tubes in oil bath | Soni et al. (2022) |
| 6.1 | |||||
| 100 | 50 | DS medium culture | Spores produced in Duncan and Strong (DS) sporulation medium | Flask technique or similar, temp fixed at 100°C, then spore suspension added, mixed and sampled at time intervals | Raju and Sarker (2007) |
| 26 | |||||
| 13 | |||||
| 95 | 2.16 | Fruit juices | Not clarified, serious shortcomings, not specified that they use anaerobiosis for incubation | Kooiman tubes technique | Brooks (2013) |
3.2.2. Salmonella Senftenberg
Data on thermal inactivation of S. Senftenberg were extracted from the review of studies on the thermal resistance of salmonellae by Doyle and Mazzotta (2000), collated in a previous EFSA scientific opinion (EFSA BIOHAZ Panel, 2021). The D values retrieved provide evidence that the physico‐chemical characteristics of the ABP or the raw materials used in the thermal processes under assessment will impact on the levels of reduction achieved. Details of the data points used are displayed in table A2 of appendix A of the previous EFSA opinion (EFSA BIOHAZ Panel, 2021).
3.2.3. Enterococcus faecalis
Data on thermal inactivation (D‐values) of E. faecalis, collated in a previous EFSA scientific opinion (EFSA BIOHAZ Panel, 2021), were used to estimate the times needed to inactivate 5 log10 units as a function of the treatment temperature. Details of the data points used are displayed in table A1 of appendix A of the former EFSA opinion (EFSA BIOHAZ Panel, 2021).
3.2.4. Viruses
A total of 11 D‐values or point estimates of the time needed to achieve 1 log reduction were retrieved through literature search for Adenoviridae (Shirasaki et al., 2020; Peter and Kühnel, 2020; Maheswari et al., 2004; Tuladhar et al., 2012; Walker et al., 1982), 4 for Anelloviridae (Welch et al., 2006), 8 for Circoviridae (O'Dea et al., 2008; Welch et al., 2006) and 26 for Parvoviridae (Spillmann et al., 1987; Brauniger et al., 2000; Blumel et al., 2002; Paluszak et al., 2010; Elving et al., 2014; Huangfu et al., 2017; Gröner et al., 2018; Gemmell et al., 2021). Further 19 values (Lelie et al., 1987; Lund et al., 1996; Yunoki et al., 2003; Sahlström et al., 2008; Nims and Plavsic, 2013; Nims and Zhou, 2016) were retrieved for Parvoviridae from a previous EFSA Opinion (EFSA BIOHAZ Panel, 2021).
The comparison of data points obtained for the four viral families, showed a higher thermal susceptibility of Adenoviridae while, based on the few values available for Anelloviridae and Circoviridae, their heat resistance was comparable or slightly lower than that of Parvoviridae, confirming the high heat resistance of this latter viral family (Nims and Plavsic, 2013). Parvoviruses were therefore selected as the most thermal resistant viral hazards for the assessment.
The information related to the thermal resistance of members of the family Parvoviridae retrieved through the extensive literature search, including the matrices and the conditions applied in the different inactivation studies, is summarised in Table B.1 of Appendix B. Time required for 3 log10 reduction of the four families of viruses at different temperatures are displayed in Figure B.1 of Appendix B.
Figure B.1.
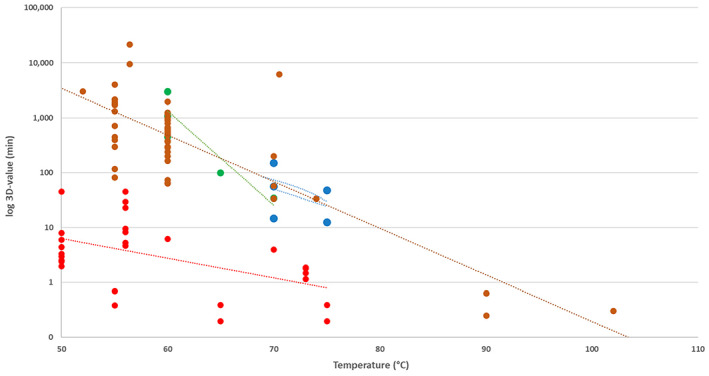
Time in minutes to achieve 3 log10 reduction of the pre‐selected families of virus parvoviruses in different matrixes (mixed liquid, whole milk, ground beef, digestion waste) and temperatures obtained from the ELS (3‐fold the estimated D value, assuming log‐linear behaviour). Lines represent the fit of the Bigelow model to the corresponding data set. Blue: Circoviridae. Green: Annelloviridae. Red: Adenoviridae. Brown: Parvoviridae
3.3. What is the ‘level of inactivation’ achieved for methods 2–5 and 7? (AQ4)
3.3.1. Methods 2–5 and 7
Table 5 compiles the heat resistance parameters (log10Dref and z) and their standard errors, which were obtained by fitting the Bigelow equation to the data retrieved for the target pathogens. According to these meta‐analytical data, C. perfringens spores presented greater heterogeneity in the relationship between log10Dref and temperature (Figure 5) than S. Senftenberg (Figure 6), E. faecalis (Figure 7) and parvoviruses (Figure 8).
Table 5.
Estimates and standard errors (SE) of the thermal inactivation parameters obtained from the data retrieved for spores of Clostridium perfringens, Salmonella Senftenberg, Enterococcus faecalis and parvoviruses used in the simulations. Correlation values between estimates and standard deviations (SD) of residuals are also shown
| Microorganism | Log10Dref | SE (log10Dref) | z | SE (z) | Correlation | SD (residuals) |
|---|---|---|---|---|---|---|
| Spores of Clostridium perfringens (Methods 2–4, 7). Data: all matrices except water Tref = 105°C (N = 89) | 0.647 | 0.128 | 19.19 | 5.549 | 0.798 | 0.729 |
|
Clostridium perfringens (Method 5) Subset: beef slurry, beef gravy, pork luncheon roll Tref = 105°C (N = 11) |
0.268 | 0.129 | 10.28 | 1.041 | 0.404 | 0.125 |
|
Salmonella Senftenberg (Methods 2–4,7) Data: all matrices Tref = 90°C (N = 53) |
−2.501 | 0.635 | 8.899 | 1.086 | 0.632 | 0.382 |
|
Salmonella Senftenberg (Method 5) Data: all matrices except chocolate and raw milk Tref = 68°C (N = 44) |
−0.777 | 0.255 | 6.465 | 0.653 | 0.649 | 0.340 |
|
Enterococcus faecalis (All Methods) Data: all matrices Tref = 72°C (N = 20) |
−0.692 | 0.217 | 7.421 | 0.667 | 0.622 | 0.263 |
|
Parvoviruses (All Methods) Data: all matrices Tref = 117°C (N = 42) |
−1.522 | 0.342 | 15.00 | 1.462 | 0.915 | 0.534 |
Figure 5.
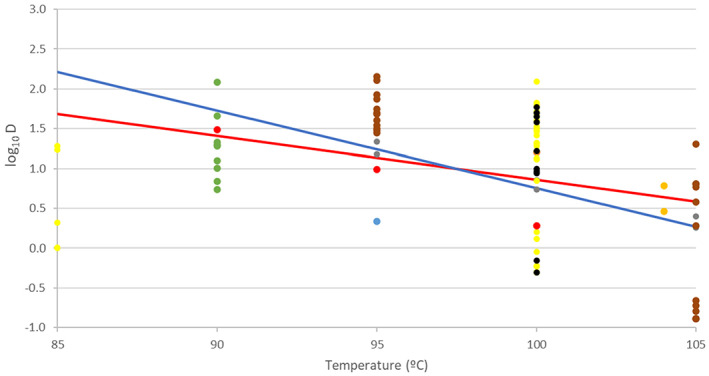
Scatter plot of the log10 D [min] of Clostridium perfringens spores in different matrices versus temperature, showing Bigelow models for use in methods 2–4 and 7 (fitted to all data; red line) and method 5 (fitted to beef slurry, beef gravy and pork luncheon roll; blue line). Black: water or broth; orange: beef gravy; yellow: DS (Duncan and Strong medium); red: pork luncheon roll; grey: beef slurry; blue: fruit juice; brown: sodium phosphate buffer; dark orange: distilled water; green: not stated
Figure 6.
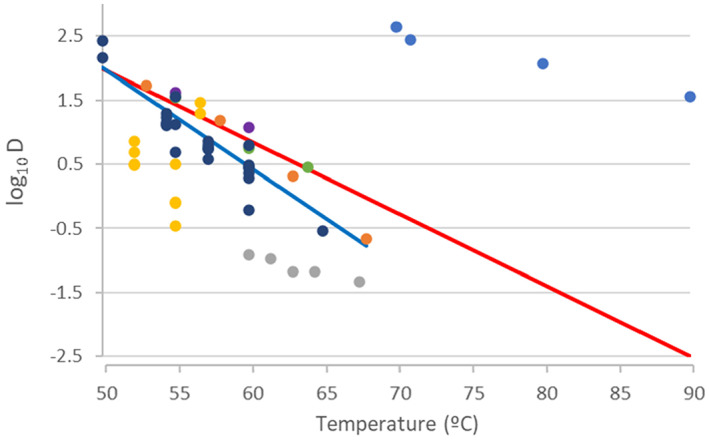
Scatter plot of the log10 D [min] of Salmonella Senftenberg in different matrices versus temperature, showing Bigelow models for use in methods 2–4 and 7 (fitted to all data; red line) and method 5 (fitted to all data except chocolate and raw milk; blue line). Blue: chocolate; orange: egg whites; grey: raw milk; dark orange: egg whites; purple: egg yolks; light green: whole eggs; black: nutrient broth
Figure 7.
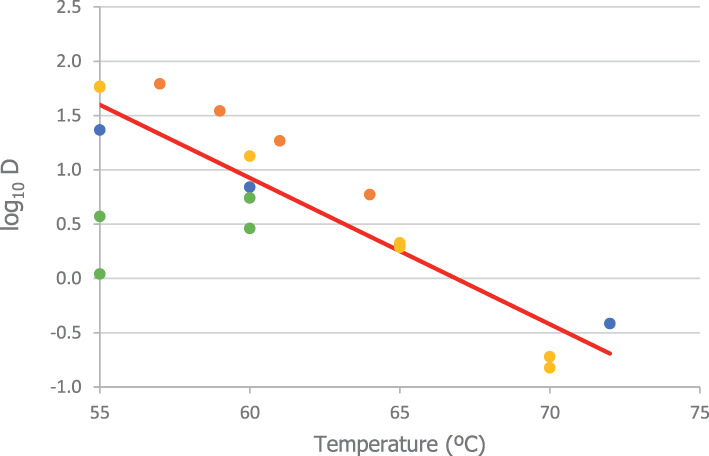
Scatter plot of the log10 D [min] of Enterococcus faecalis in different matrices versus temperature, showing the fitted Bigelow model for use in all methods. Blue: mixed liquid; orange: ground beef; dark orange: whole milk; green: digestion waste
Figure 8.
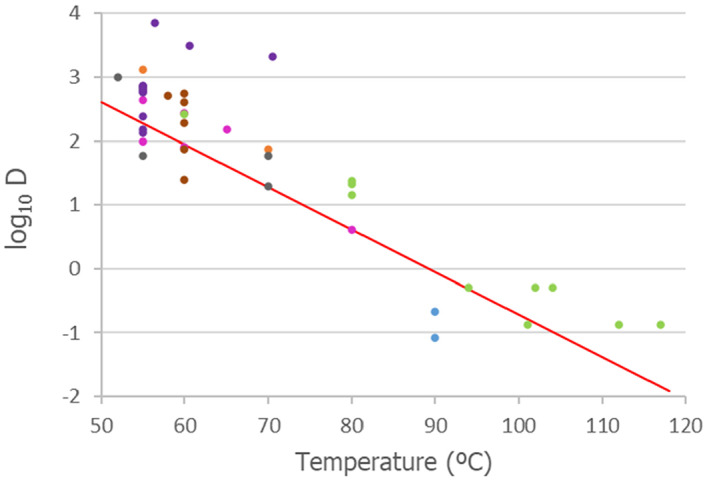
Scatter plot of the log10 D [min] of parvoviruses in different matrices versus temperature, showing the fitted Bigelow model for use in all methods. Pink: culture medium; blue: glucose concentrate; green: water; dark orange: saline solution; brown: plasma derivatives; purple: virus on filters; green: water; dark green: semi‐solid medium
These graphs also show the fitted lines corresponding to the Bigelow model. In the cases of C. perfringens spores and S. Senftenberg, two model solutions were achieved and are presented in the graph: the red line for use in simulations pertaining to methods 2–4 and 7, and the blue line for the simulations pertaining to method 5. The red line represents a solution for data from studies reporting the highest thermal resistance in combination with matrices of a composition resembling that of processed animal protein. The blue line is a solution for matrices that are low in fat and high in protein. For both microorganisms, spores of C. perfringens and S. Senftenberg, the models combining matrices low in fat and high in protein provided lower log10Dref and z estimates. This means that in proteinaceous matrices, both microorganisms are less resistant to heat. For both E. faecalis and parvoviruses only one Bigelow model solution was attained since the low number and the nature of the matrices recovered did not make such an assessment possible. For easier comparison of the thermal resistance between the target pathogens, the mean predictions of the six Bigelow models fitted are shown in Figure 9. According to the log‐linear inactivation data recovered, greater resistance in response to temperature increase is displayed by spores of C. perfringens followed by parvoviruses (lower slopes in Figure 9).
Figure 9.
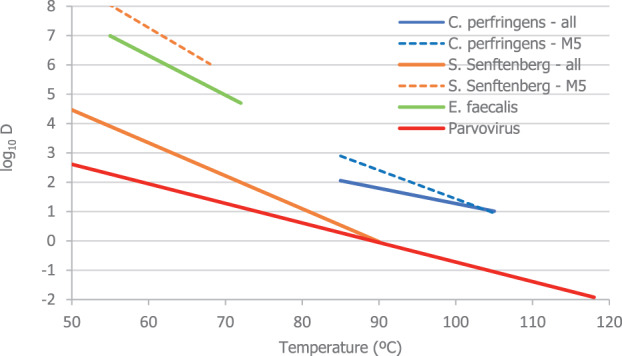
Predicted lines of the Bigelow models describing log10 D [min] as a function of temperature for the four target pathogens. Thick lines denote models fitted to all available matrices, and dashed lines to proteinaceous matrices. The temperature range of the lines covers the range for which data were available, to prevent extrapolation
Table 6 summarises the probabilities of achieving the target reduction of selected bacterial and viral indicators when Methods 2 to 5, in both the coincidental and consecutive modes, and the five scenarios for Method 7, are applied.
Table 6.
Probability to achieve the target inactivation standards for spores of Clostridium perfringens, Salmonella Senftenberg, Enterococcus faecalis and parvoviruses according to the results of the model
| Method | Indicators | Target log10 reduction |
Probability to achieve at least the target log10 reduction Mode: coincidental |
Probability to achieve at least the target log10 reduction Mode: consecutive |
Probability to achieve at least the target log10 reduction |
|---|---|---|---|---|---|
| 2 | Spores of C. perfringens | 5 | 1.0000 | 1.0000 | |
| S. Senftenberg | 5 | 1.0000 | 1.0000 | ||
| E. faecalis | 5 | 1.0000 | 1.0000 | ||
| Parvoviruses | 3 | 1.0000 | 1.0000 | ||
| 3 | Spores of C. perfringens | 5 | 1.0000 | 1.0000 | |
| S. Senftenberg | 5 | 1.0000 | 1.0000 | ||
| E. faecalis | 5 | 1.0000 | 1.0000 | ||
| Parvoviruses | 3 | 1.0000 | 1.0000 | ||
| 4 | Spores of C. perfringens | 5 | 0.0660 | 0.9240 | |
| S. Senftenberg | 5 | 1.0000 | 1.0000 | ||
| E. faecalis | 5 | 1.0000 | 1.0000 | ||
| Parvoviruses | 3 | 0.9963 | 0.9998 | ||
| 5 | Spores of C. perfringens | 5 | 0.9970 | 0.9970 | |
| S. Senftenberg | 5 | 1.0000 | 1.0000 | ||
| E. faecalis | 5 | 1.0000 | 1.0000 | ||
| Parvoviruses | 3 | 0.9984 | 0.9992 | ||
| 7 scenario 1 | Spores of C. perfringens | 5 | 0.0000 | ||
| S. Senftenberg | 5 | 0.9966 | |||
| E. faecalis | 5 | 1.0000 | |||
| Parvoviruses | 3 | 0.0735 | |||
| 7 scenario 2 | Spores of C. perfringens | 5 | 0.6850 | ||
| S. Senftenberg | 5 | 1.0000 | |||
| E. faecalis | 5 | 1.0000 | |||
| Parvoviruses | 3 | 0.9950 | |||
| 7 scenario 3 | Spores of C. perfringens | 5 | 0.9999 | ||
| S. Senftenberg | 5 | 1.0000 | |||
| E. faecalis | 5 | 1.0000 | |||
| Parvoviruses | 3 | 1.0000 | |||
| 7 scenario 4 | Spores of C. perfringens | 5 | 0.0040 | ||
| S. Senftenberg | 5 | 1.0000 | |||
| E. faecalis | 5 | 1.0000 | |||
| Parvoviruses | 3 | 0.9905 | |||
| 7 scenario 5 | Spores of C. perfringens | 5 | 0.0000 | ||
| S. Senftenberg | 5 | 0.9999 | |||
| E. faecalis | 5 | 0.9993 | |||
| Parvoviruses | 3 | 0.9256 |
The results of the model show that for the four selected indicators, the lowest probability of achieving the target level of inactivation in all methods was estimated for the spores of C. perfringens. Thus, if the model predicts a high probability of 5 log10 inactivation of C. perfringens, the probability will be even higher for the other indicators, becoming more likely to achieve the required standard of 5 log10 or 3 log10.
The results of the model showed a probability of inactivation of at least 5 log10 of spores of C. perfringens over 0.99 of the iterations, for methods 2, 3 and 5, in both the coincidental and consecutive modes and 0.92 for method 4 in consecutive mode. For method 4 in coincidental mode, the model estimated a probability of 0.066. For method 7, the model estimates a probability of 0.004 or below of achieving the 5 log10 reduction for scenarios 1, 4 and 5, and probabilities of 0.685 and 0.999 for achieving the same level of reduction for scenarios 2 and 3, respectively.
Table 7 shows the outcomes of the simulations undertaken to determine the probability of achieving specific log10 reductions against spores of C. perfringens considering different log reduction levels ranging between < 5 log10 and > 12 log10.
Table 7.
Probability of achieving specific log10 reductions in different ranges, and expected value of log10 reduction (in brackets) of spores of Clostridium perfringens after application of coincidental and consecutive heat treatments
| Method | Log10 reduction |
Probability of reduction in coincidental mode (Expected value of log10 reduction) |
Probability of reduction in consecutive mode (Expected value of log10 reduction) |
Probability of reduction (Expected value of log10 reduction) |
|---|---|---|---|---|
| 2 |
L < 5 5 ≤ L < 6 6 ≤ L < 8 8 ≤ L < 10 10 ≤ L < 12 L ≥ 12 |
0.000 0.000 0.000 0.000 0.002 0.998 |
0.000 0.000 0.000 0.000 0.000 1.000 |
|
| (27.6) | (53.8) | |||
| 3 |
L < 5 5 ≤ L < 6 6 ≤ L < 8 8 ≤ L < 10 10 ≤ L < 12 L ≥ 12 |
0.000 0.000 0.002 0.019 0.065 0.914 |
0.000 0.000 0.000 0.000 0.001 0.999 |
|
| (17.3) | (27.1) | |||
| 4 |
L < 5 5 ≤ L < 6 6 ≤ L < 8 8 ≤ L < 10 10 ≤ L < 12 L ≥ 12 |
0.934 0.051 0.014 0.001 0.000 0.000 |
0.076 0.147 0.408 0.257 0.086 0.026 |
|
| (3.30) | (7.39) | |||
| 5 |
L < 5 5 ≤ L < 6 6 ≤ L < 8 8 ≤ L < 10 10 ≤ L < 12 L ≥ 12 |
0.003 0.016 0.136 0.265 0.260 0.320 |
0.003 0.015 0.127 0.256 0.265 0.334 |
|
| (10.7) | (10.8) | |||
| 7, Scenario 1 |
L < 5 5 ≤ L < 6 6 ≤ L < 8 8 ≤ L < 10 10 ≤ L < 12 L ≥ 12 |
1.000 0.000 0.000 0.000 0.000 0.000 |
||
| (0.157) | ||||
| 7, Scenario 2 |
L < 5 5 ≤ L < 6 6 ≤ L < 8 8 ≤ L < 10 10 ≤ L < 12 L ≥ 12 |
0.315 0.252 0.345 0.078 0.009 0.001 |
||
| (6.1) | ||||
| 7, Scenario 3 |
L < 5 5 ≤ L < 6 6 ≤ L < 8 8 ≤ L < 10 10 ≤ L < 12 L ≥ 12 |
0.001 0.005 0.055 0.154 0.217 0.568 |
||
| (12.6) | ||||
| 7, Scenario 4 |
L < 5 5 ≤ L < 6 6 ≤ L < 8 8 ≤ L < 10 10 ≤ L < 12 L ≥ 12 |
0.996 0.003 0.001 0.000 0.000 0.000 |
||
| (2.25) | ||||
| 7, Scenario 5 |
L < 5 5 ≤ L < 6 6 ≤ L < 8 8 ≤ L < 10 10 ≤ L < 12 L ≥ 12 |
1.000 0.000 0.000 0.000 0.000 0.000 |
||
| (1.12) |
The log10 reduction of C. perfringens spores achieved by the inactivation methods are also illustrated as cumulative distributions in Figures 10 and 11. The left skewness of these distributions (i.e. the further it lies to the right) is linked to a greater level of inactivation. The decreasing order of the estimated lethality is: method 2 > method 3 > method 5 > method 4. The probability of insufficient inactivation (lower than 5 log10) is associated with the left tail. In Figure 10, the vertical threshold of 5 log10 cycles was placed on the graph for comparison with the target reduction.
Figure 10.
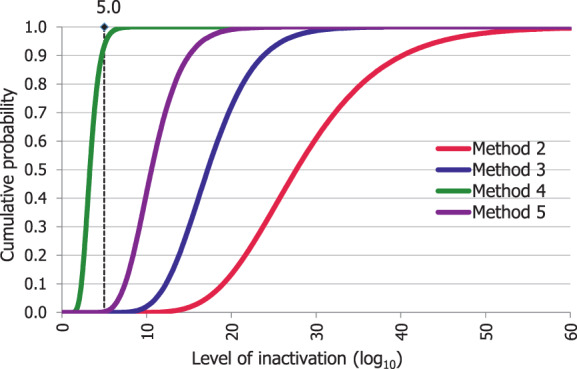
Cumulative probability of the lethality of the heat treatment methods 2, 3, 4 and 5 in coincidental mode against Clostridium perfringens spores
Figure 11.
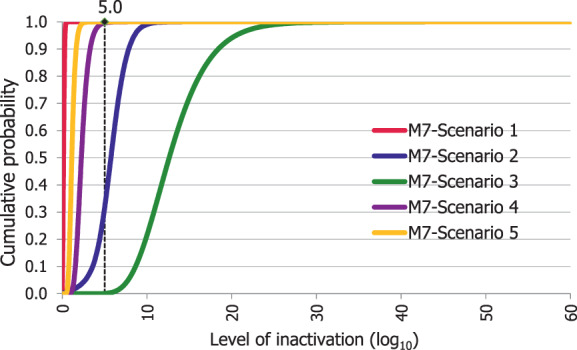
Cumulative probability of the lethality of the five scenarios of heat treatment method 7 against Clostridium perfringens spores
For method 7, the mildness of the heat regimes of scenarios 1 and 5 is evident in their respective lethality cumulative distributions in Figure 11. The latter shows that the lethality against C. perfringens spores is higher in scenario 3, followed by scenario 2, scenario 4, scenario 5 and scenario 1, being the last three scenarios on the left side of the threshold.
Tables C.1, C.2 and C.3 of Appendix C show the outcomes of the simulations undertaken to determine the probability of achieving specific log10 reductions against S. Senftenberg, E. faecalis and parvoviruses considering different log reduction levels, ranging between < 5 log10 (< 3 log10 for parvoviruses) to > 12 log10. The log10 reduction of parvoviruses achieved by the inactivation methods are also illustrated as cumulative distributions in Figures C.1 and C.2.
Table C.1.
Probability of achieving specific log10 reductions in different ranges, and expected value of log10 reduction (in brackets) of Salmonella Senftenberg after application of coincidental and consecutive heat treatments
| Method | Log10 reduction | Probability of reduction in coincidental mode (Expected value of log10 reduction) | Probability of reduction in consecutive mode (Expected value of log10 reduction) | Probability of reduction (Expected value of log10 reduction) |
|---|---|---|---|---|
| 2 |
L < 5 5 ≤ L < 6 6 ≤ L < 8 8 ≤ L < 10 10 ≤ L < 12 L ≥ 12 |
0.000 0.000 0.000 0.000 0.000 1.000 |
0.000 0.000 0.000 0.000 0.000 1.000 |
|
| (39,620) | (93,502) | |||
| 3 |
L < 5 5 ≤ L < 6 6 ≤ L < 8 8 ≤ L < 10 10 ≤ L < 12 L ≥ 12 |
0.000 0.000 0.000 0.000 0.000 1.000 |
0.000 0.000 0.000 0.000 0.000 1.000 |
|
| (30,110) | (51,664) | |||
| 4 |
L < 5 5 ≤ L < 6 6 ≤ L < 8 8 ≤ L < 10 10 ≤ L < 12 L ≥ 12 |
0.000 0.000 0.000 0.000 0.001 0.999 |
0.000 0.000 0.000 0.000 0.000 1.000 |
|
| (5,071) | (12,678) | |||
| 5 |
L < 5 5 ≤ L < 6 6 ≤ L < 8 8 ≤ L < 10 10 ≤ L < 12 L ≥ 12 |
0.000 0.000 0.000 0.000 0.000 1.000 |
0.000 0.000 0.000 0.000 0.000 1.000 |
|
| (718) | (1,077) | |||
| 7, Scenario 1 |
L < 5 5 ≤ L < 6 6 ≤ L < 8 8 ≤ L < 10 10 ≤ L < 12 L ≥ 12 |
0.000 0.001 0.001 0.002 0.002 0.994 |
||
| (333) | ||||
| 7, Scenario 2 |
L < 5 5 ≤ L < 6 6 ≤ L < 8 8 ≤ L < 10 10 ≤ L < 12 L ≥ 12 |
0.000 0.000 0.000 0.000 0.000 1.000 |
||
| (28,526) | ||||
| 7, Scenario 3 |
L < 5 5 ≤ L < 6 6 ≤ L < 8 8 ≤ L < 10 10 ≤ L < 12 L ≥ 12 |
0.000 0.000 0.000 0.000 0.000 1.000 |
||
| (17,750) | ||||
| 7, Scenario 4 |
L < 5 5 ≤ L < 6 6 ≤ L < 8 8 ≤ L < 10 10 ≤ L < 12 L ≥ 12 |
0.000 0.000 0.000 0.000 0.001 0.999 |
||
| (3,169) | ||||
| 7, Scenario 5 |
L < 5 5 ≤ L < 6 6 ≤ L < 8 8 ≤ L < 10 10 ≤ L < 12 L ≥ 12 |
0.000 0.000 0.000 0.000 0.001 0.999 |
||
| (1,585) |
Table C.2.
Probability of achieving specific log10 reductions in different ranges, and expected value of log10 reduction (in brackets) of Enterococcus faecalis after application of coincidental and consecutive heat treatments
| Method | Log10 reduction | Probability of reduction in coincidental mode (Expected value of log10 reduction) | Probability of reduction in consecutive mode (Expected value of log10 reduction) | Probability of reduction (Expected value of log10 reduction) |
|---|---|---|---|---|
| 2 |
L < 5 5 ≤ L < 6 6 ≤ L < 8 8 ≤ L < 10 10 ≤ L < 12 L ≥ 12 |
0.000 0.000 0.000 0.000 0.000 1.000 |
0.000 0.000 0.000 0.000 0.000 1.000 |
|
| (615) | (1,452) | |||
| 3 |
L < 5 5 ≤ L < 6 6 ≤ L < 8 8 ≤ L < 10 10 ≤ L < 12 L ≥ 12 |
0.000 0.000 0.000 0.000 0.000 1.000 |
0.000 0.000 0.000 0.000 0.000 1.000 |
|
| (467) | (802) | |||
| 4 |
L < 5 5 ≤ L < 6 6 ≤ L < 8 8 ≤ L < 10 10 ≤ L < 12 L ≥ 12 |
0.000 0.000 0.000 0.000 0.001 0.999 |
0.000 0.000 0.000 0.000 0.000 1.000 |
|
| (78.8) | (197) | |||
| 5 |
L < 5 5 ≤ L < 6 6 ≤ L < 8 8 ≤ L < 10 10 ≤ L < 12 L ≥ 12 |
0.000 0.000 0.000 0.000 0.000 1.000 |
0.000 0.000 0.000 0.000 0.000 1.000 |
|
| (590) | (886) | |||
| 7, Scenario 1 |
L < 5 5 ≤ L < 6 6 ≤ L < 8 8 ≤ L < 10 10 ≤ L < 12 L ≥ 12 |
0.000 0.000 0.000 0.000 0.001 0.999 |
||
| (68.9) | ||||
| 7, Scenario 2 |
L < 5 5 ≤ L < 6 6 ≤ L < 8 8 ≤ L < 10 10 ≤ L < 12 L ≥ 12 |
0.000 0.000 0.000 0.000 0.000 1.000 |
||
| (443) | ||||
| 7, Scenario 3 |
L < 5 5 ≤ L < 6 6 ≤ L < 8 8 ≤ L < 10 10 ≤ L < 12 L ≥ 12 |
0.000 0.000 0.000 0.000 0.000 1.000 |
||
| (275) | ||||
| 7, Scenario 4 |
L < 5 5 ≤ L < 6 6 ≤ L < 8 8 ≤ L < 10 10 ≤ L < 12 L ≥ 12 |
0.000 0.000 0.000 0.001 0.002 0.997 |
||
| (49.2) | ||||
| 7, Scenario 5 |
L < 5 5 ≤ L < 6 6 ≤ L < 8 8 ≤ L < 10 10 ≤ L < 12 L ≥ 12 |
0.001 0.002 0.010 0.024 0.040 0.924 |
||
| (24.6) |
Table C.3.
Probability of achieving specific log10 reductions in different ranges, and expected value of log10 reduction (in brackets) of Parvovirus after application of coincidental and consecutive heat treatments
| Method | Log10 reduction | Probability of reduction in coincidental mode (Expected value of log10 reduction) | Probability of reduction in consecutive mode (Expected value of log10 reduction) | Probability of reduction (Expected value of log10 reduction) |
|---|---|---|---|---|
| 2 |
L < 3 3 ≤ L < 5 5 ≤ L < 6 6 ≤ L < 8 8 ≤ L < 10 10 ≤ L < 12 L ≥ 12 |
0.000 0.000 0.000 0.000 0.000 0.000 1.000 |
0.000 0.000 0.000 0.000 0.000 0.000 1.000 |
|
| (2,470) | (3,332) | |||
| 3 |
L < 3 3 ≤ L < 5 5 ≤ L < 6 6 ≤ L < 8 8 ≤ L < 10 10 ≤ L < 12 L ≥ 12 |
0.000 0.000 0.000 0.000 0.000 0.000 1.000 |
0.000 0.000 0.000 0.000 0.000 0.000 1.000 |
|
| (1,007) | (1,290) | |||
| 4 |
L < 3 3 ≤ L < 5 5 ≤ L < 6 6 ≤ L < 8 8 ≤ L < 10 10 ≤ L < 12 L ≥ 12 |
0.000 0.000 0.000 0.000 0.000 0.000 1.000 |
0.000 0.000 0.000 0.000 0.000 0.000 1.000 |
|
| (330) | (553) | |||
| 5 |
L < 3 3 ≤ L < 5 5 ≤ L < 6 6 ≤ L < 8 8 ≤ L < 10 10 ≤ L < 12 L ≥ 12 |
0.000 0.000 0.000 0.000 0.000 0.000 1.000 |
0.000 0.000 0.000 0.000 0.000 0.000 1.000 |
|
| (154) | (160) | |||
| 7, Scenario 1 |
L < 3 3 ≤ L < 5 5 ≤ L < 6 6 ≤ L < 8 8 ≤ L < 10 10 ≤ L < 12 L ≥ 12 |
0.978 0.022 0.001 0.000 0.000 0.000 0.000 |
||
| (1,592) | ||||
| 7, Scenario 2 |
L < 3 3 ≤ L < 5 5 ≤ L < 6 6 ≤ L < 8 8 ≤ L < 10 10 ≤ L < 12 L ≥ 12 |
0.000 0.000 0.000 0.000 0.000 0.001 0.999 |
||
| (102) | ||||
| 7, Scenario 3 |
L < 3 3 ≤ L < 5 5 ≤ L < 6 6 ≤ L < 8 8 ≤ L < 10 10 ≤ L < 12 L ≥ 12 |
0.000 0.000 0.000 0.000 0.000 0.000 1.000 |
||
| (1,370) | ||||
| 7, Scenario 4 |
L < 3 3 ≤ L < 5 5 ≤ L < 6 6 ≤ L < 8 8 ≤ L < 10 10 ≤ L < 12 L ≥ 12 |
0.000 0.000 0.000 0.000 0.000 0.000 1.000 |
||
| (332) | ||||
| 7, Scenario 5 |
L < 3 3 ≤ L < 5 5 ≤ L < 6 6 ≤ L < 8 8 ≤ L < 10 10 ≤ L < 12 L ≥ 12 |
0.000 0.000 0.000 0.000 0.000 0.001 0.999 |
||
| (166) |
Figure C.1.
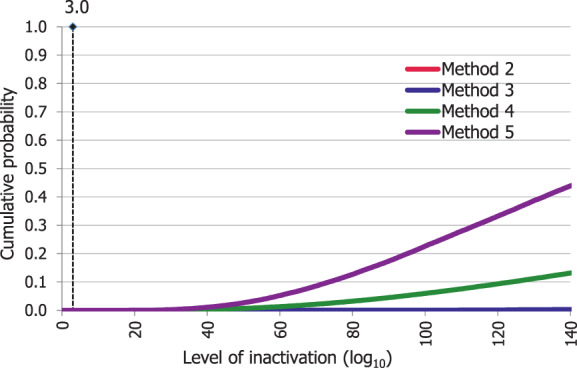
Cumulative probability of the lethality of the heat treatment methods 2, 3, 4 and 5 in coincidental mode against parvoviruses
Figure C.2.
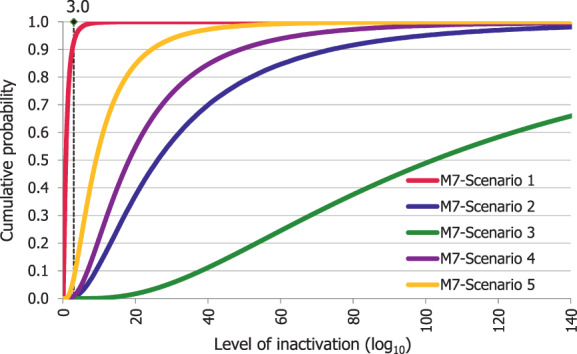
Cumulative probability of the lethality of the five scenarios of heat treatment method 7 (M7) against parvoviruses
3.3.2. Comparative analysis with Clostridium botulinum
As explained in Section 2.3.4, in previous EFSA ABP scientific opinions, extrapolation at higher temperatures than the experimental ones was discouraged as an approach for estimating the lethality of alternative methods. Therefore, for some of the methods under assessment, the reductions that can be reached at the actual treatment temperatures of the heat regime may be underestimated.
In Section 3.1.1.1, criteria for the selection of relevant bacterial pathogens, it is highlighted that in previous EFSA assessments of alternative ABP processing methods applied to Category 3 ABP, if the hazard identification considers spore‐forming bacteria as relevant biological hazards, a requirement of 12 log10 reduction in C. botulinum spores, equivalent to a treatment at 121.1°C for 3 min, can be considered also effective to achieve a 5 log10 reduction of other less heat resistant spore‐forming bacteria, such as C. perfringens. The equivalent accumulated lethality expressed in minutes at Tref 121°C of methods 2 to 5 and the scenarios of method 7 considered in this opinion at temperatures other than the reference one for C. botulinum (121.1°C) was calculated considering a z‐value of 10°C. The values reached would be:
Method 2:38.81 min
Method 3:10.09 min
Method 4: 27.16 min
Method 5:0.46 min
Method 7 scenario 1:0.001 min
Method 7 scenario 2:0.22 min
Method 7 scenario 3: 13.74 min
Method 7 scenario 4:24.54 min
Method 7 scenario 5:77.44 min
Therefore, all methods assessed, except method 5 and scenarios 1 and 2 of method 7, would exceed the requirement for C. botulinum (equivalent to > 3 min at 121°C in terms of lethality). Consequently, it would be expected that they will achieve at least a 5 log10 reduction of other less heat resistant bacterial spores, such as those of C. perfringens. Considering the outputs of the model, method 4 coincidental and method 7 scenarios 4 and 5 would have a low probability of achieving the 5 log10 reduction of C. perfringens, but they would achieve the requirements for C. botulinum (12 log reduction). This is due to the fact that extrapolation of the model at temperatures above 105°C for C. perfringens is not applied, while method 4 and scenarios 4 and 5 of method 7 involve treatments at much higher temperatures (in the range of 125–133°C). In these cases, the model is very likely underestimating the lethality achieved by the processing methods, and the fact that the methods are predicted to reach a sufficient level of reduction (> 12 log10) of spores of C. botulinum provides an indication that a sufficient level of reduction of the less heat resistant C. perfringens spores would be also achieved.
The opposite happens for method 5, for which the model predicts a probability of over 99% of achieving the 5 log10 reduction of C. perfringens, whereas the requirement for C. botulinum is not met (equivalent to < 3 min at 121°C in terms of lethality). In this case, this indicates that the process is possibly not achieving a 12 log10 reduction of C. botulinum spores, but it is producing a 5 log10 reduction of spores of C. perfringens, identified as the relevant hazard to be considered in the assessment.
In scenarios 1 and 2 of method 7, the estimations show that the minimum level of inactivation required is possibly not achieved either for C. perfringens (5 log10 reduction), or for C. botulinum (12 log10 reduction). In these cases, the hygienic conditions and the nature of the material will ultimately determine the safety of the end product, as the effectiveness of those heat regimes is limited. It is important to highlight that the material treated by method 7 is subject to the safety/hygiene verification by the microbiological standards established in Chapter III, Annex IV of Commission Regulation (EU) 142/2011.
3.4. AQ5: What is the certainty that the ‘level of inactivation’ achieved by methods 2–3–4‐5 and 7 as in AQ4 is sufficient to reach the standards for category 3 ABP?
The following EKE question was used to address AQ5: What is the probability that a 5 log10 reduction of spores of C. perfringens is achieved, in more than 99% of the cases, by application of each of the relevant processes (methods 2, 3, 4, 5 in coincidental mode and five t/T combinations of method 7), assuming that the processes are performed as prescribed and that the indicated process conditions are achieved?
The reason why only the spores C. perfringens are considered in the EKE question is because (according to the results of this assessment) they represent the most thermal resistant microorganism among the selected indicators. The coincidental mode of the methods 2 to 5 was assessed as it has been considered in this assessment as the worst‐case scenario.
The results of the consensus judgements for each of the t/T combinations are given in Table 8 and Figure 12. Based on the results, a 5 log10 reduction of C. perfringens spores was judged: 99–100% certain for methods 2 and 3 in coincidental mode; 98–100% certain for method 7 scenario 3; 80–99% certain for method 5 in coincidental mode; 66–100% certain for method 4 in coincidental mode and for method 7 scenarios 4 and 5; 25–75% certain for method 7 scenario 2; and 0–5% certain for method 7 scenario 1. The same or higher certainty to achieve the 5 log10 reduction of C. perfringens spores is expected when methods 2 to 5 are applied in consecutive mode.
Table 8.
Time/temperature (t/T) combinations for each or the processes (methods 2, 3, 4, 5 in coincidental mode and five t/T combinations of method 7) included in the EKE and results of the consensus judgement
| Process | Combination t/T | Consensus judgement | |||
|---|---|---|---|---|---|
| Method 2 – coincidental mode | 100°C × 5′ | 110°C × 70′ | 120°C × 50′ | – | 99–100% |
| Method 3 – coincidental | 100°C × 40′ | 110°C × 42′ | 120°C × 13′ | – | 99–100% |
| Method 4 – coincidental | 100°C × 3′ | 110°C × 5′ | 120°C × 5′ | 130°C × 3′ | 66–100% |
| Method 5 – coincidental | 80°C × 60′ | 100°C × 60′ | – | – | 80–99% |
| Method 7 – scenario 1 | 80°C × 14′ | 0–5% | |||
| Method 7 – scenario 2 | 95°C × 90′ | 25–75% | |||
| Method 7 – scenario 3 | 115°C × 56′ | 98–100% | |||
| Method 7 – scenario 4 | 125°C × 10′ | 66–100% | |||
| Method 7 – scenario 5 | 133°C × 5′ | 66–100% | |||
Figure 12.
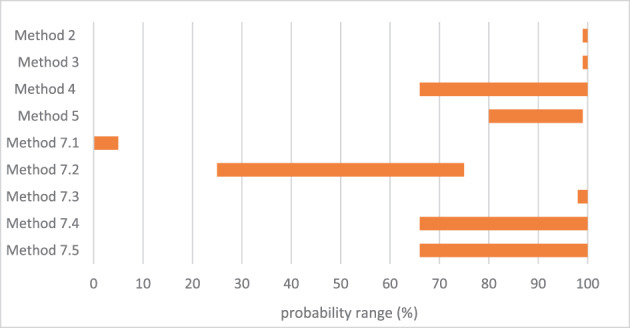
Probability ranges obtained in the EKE, indicating how certain the experts are that a 5 log10 reduction of spores of Clostridium perfringens is achieved, in more than 99% of the cases, by application of each of the relevant processes (methods 2, 3, 4, 5 in coincidental mode and five t/T combinations of method 7), assuming that the processes are performed as prescribed and that the indicated process conditions are achieved
3.5. Uncertainty analysis
4. Conclusions
Four relevant pathogens were selected as indicators to assess the efficacy of standard processing methods 2, 3, 4, 5 and 7 for Category 3 ABP of porcine origin: S. Senftenberg, E. faecalis, spores of C. perfringens and parvoviruses. It was agreed to assess whether the methods would reach a level of reduction of 5 log10 for the bacterial indicators and 3 log10 for the viral indicator. This approach has been considered a worst‐case scenario, as Category 3 ABP are only sourced from animals slaughtered fit for human consumption or rejected but not showing signs of disease communicable to humans or animals.
The time/temperature (t/T) parameters of processing methods 2, 3, 4 and 5 are stated in Chapter III, Annex IV of Commission Regulation (EU) 142/2011: method 2–100°C for at least 125 min, 110°C for at least 120 min and 120°C for at least 50 min; method 3–100°C for at least 95 min, 110°C for at least 55 min and 120°C for at least 13 min; method 4–100°C for at least 16 min, 110°C for at least 13 min, 120°C for at least 8 min and 130°C for at least 3 min; method 5–80°C for at least 120 min and 100°C for at least 60 min. The efficacy of these processing methods was assessed either when the core temperatures are achieved consecutively or through a coincidental combination of the time periods indicated. The latter has been considered as the worst‐case scenario.
For method 7, the legislation does not state time/temperature parameters. However, there are process parameters approved in different EU member states based on equipment used and treated raw materials. Based on the approved parameters provided by the industry, five scenarios of individual time/temperature profiles were selected for the scope of this assessment in order to apply for method 7 the same methodological approach followed for methods 2 to 5. The selected scenarios were: scenario 1–80°C for at least 14 min; scenario 2–95°C for at least 90 min; scenario 3–115°C for at least 56 min; scenario 4–125°C for at least 10 min; scenario 5–133°C for at least 5 min.
The level of inactivation of the selected indicators was calculated using the parameters of the Bigelow model fitted to thermal inactivation parameters retrieved for the four indicators from a previous EFSA scientific opinion (EFSA BIOHAZ Panel, 2021) and/or from extensive literature searches. Matrices with different levels of fat and proteins were included in the data modelling.
The model to estimate the level of inactivation of relevant pathogens was applied to limited experimental data and built up with certain assumptions that may result in an under‐estimation of the levels of reduction achieved, mostly for methods for which the temperature is above those for which thermal inactivation data are available.
Considering the data retrieved from the literature and the model estimates for the four selected indicators, the lowest probability of achieving the target level of inactivation was estimated for the spores of C. perfringens for all methods.
The results of the simulation showed a probability of inactivation of at least 5 log10 of spores of C. perfringens of over 0.99 for methods 2, 3 and 5, in both coincidental and consecutive modes and 0.92 for method 4 in consecutive mode. For method 4 in coincidental mode, the model estimated a probability of 0.066. For method 7, the model estimated a probability of 0.004 or below of achieving the 5 log10 reduction for scenarios 1, 4 and 5, and of 0.685 and 0.999 for achieving the same level of reduction for scenarios 2 and 3, respectively. The calculation of such probabilities was driven by the uncertainty about the thermal inactivation parameters of the target microorganisms, which could be attributed to the different strains assessed in the primary studies, the matrices and the experimental design.
An expert knowledge elicitation (EKE) was conducted to elucidate what the probability is that a 5 log10 reduction of C. perfringens spores is achieved, in more than 99% of cases, by application of the relevant processes (methods 2, 3, 4, 5 in coincidental mode and five t/T combinations of method 7) assuming that the processes are performed as prescribed and that the indicated process conditions are achieved. The information considered in the expert judgements was based on the information and data available in the draft opinion at the time of conducting the EKE, also considering evidence on inactivation of spores of C. botulinum as an additional surrogate. The latter is related to the underestimation of the model since the extrapolation at temperatures above 105°C for C. perfringens was not applied, while method 4 and scenarios 4 and 5 of method 7 involve treatments at much higher temperatures.
-
Based on the EKE, the certainty of achieving a 5 log10 reduction of C. perfringens spores (which also assure the target inactivation for the other relevant pathogens) was:
99–100% certain for methods 2 and 3 in coincidental mode; 98–100% certain for method 7 scenario 3;
80–99% certain for method 5 in coincidental mode;
66–100% certain for method 4 in coincidental mode and for method 7 scenarios 4 and 5;
25–75% certain for method 7 scenario 2; and
0–5% certain for method 7 scenario 1.
Compared to the results of the EKE for methods 2–5 in coincidental mode, the same or higher certainty to achieve the 5 log10 reduction of C. perfringens spores is expected when methods 2 to 5 are applied in consecutive mode.
Abbreviations
- ABP
Animal by‐products
- AHAW
Animal Health and Animal Welfare
- AHL
Animal Health Law
- AQ
Assessment question
- ASF
African Swine Fever
- AstV
Astrovirus
- BIOHAZ
biological hazards
- BPV
bovine parvovirus
- CPV
canine parvovirus
- CSF
classical swine fever
- DS
Duncan Strong
- D value
The time (in minutes) of exposure at a given temperature that causes a one‐log10 or 90% reduction in the population of a specific microorganism
- ECDC
European Centre for Disease Control
- EFPRA
European Fat processors and Renderers Association
- EKE
Expert Knowledge Elicitation
- ELS
extensive literature search
- EMCV
encephalomyocarditis virus
- F
The time necessary to destroy a given number of microorganisms at a reference temperature, usually 121°C for spores or 60°C for vegetative cells
- FMDV
Foot and Mouth disease virus
- GARV
Rotavirus group A
- HBV
Hepatitis B virus
- HEV
Hepatitis E virus
- L
lethality
- MVM
Minute virus of mice
- NOV
European swine Norovirus
- PAP
Processed animal protein
- PAdV
Porcine adenovirus
- PBo‐likeV
Porcine boca‐like virus
- PboV
Porcine bocavirus
- PCAD
Porcine adenovirus
- PCV
Porcine circovirus
- PCMV
Porcine cytomegalovirus
- PDCoV
Porcine deltacoronavirus
- PDNS
Porcine dermatitis and nephropathy syndrome
- PEC
Porcine enteric calicivirus
- PMWS
Post‐weaning multisystemic wasting syndrome
- PPIV
Porcine parainfluenza virus
- PPV
Porcine parvovirus
- PRRS
Porcine respiratory and reproductive syndrome
- SD
Standard deviation
- SE
Standard error
- SIV
Swine influenza virus
- SQ
Sub‐assessment question
- t/T
Time/temperature
- TTSuV
Torque Teno virus
- WOAH
Organization of Animal Health and Welfare
- z
The number of degrees the temperature has to be increased to achieve a tenfold (i.e. 1 log10) reduction in the D‐value
Appendix A – Data extracted on thermal inactivation parameters of spores of Clostridium perfringens
1.
Appendix B – Data extracted on thermal inactivation parameters of the family Parvoviridae
1.
Figure B.1 Shows the time required for 3 log10 reduction of the four families of viruses at different temperatures.
Appendix C – Results of the model for other indicators
1.
The estimation of the level of inactivation and probabilities of inactivation by the heat treatments against S. Senftenberg as estimated by the model are displayed in Table C.1. Despite the most conservative approach applied by the model, the results showed a probability of inactivation of at least 5 log10 of 100% for all methods and implementation modes.
The estimation of the lethality and probabilities of inactivation by the heat treatments against E. faecalis as estimated by the model are displayed in Table C.2. Despite the most conservative approach applied by the model, the results showed a probability of 0.99 or above of an inactivation of E. faecalis equal or greater than 5 log10.
The estimation of the lethality and probabilities of inactivation by the heat treatments against Parvovirus as estimated by the model are displayed in Table C.3. All combinations of methods and types of application have a probability of 0.99 or above to achieve an inactivation greater than 3 log10 except for method 7 scenario 1 having a probability of 0.023 of achieving an inactivation greater than 3 log10.
The log10 reduction of parvoviruses achieved by the inactivation methods are also illustrated as cumulative distributions in Figures C.1 and C.2. In this case, the lethality of the methods has the decreasing order method 2 > method 3 > method 4 > method 5. The probability of insufficient inactivation (lower than 3 log10) is associated with the left tail and when the expected value of the distribution is high (as is the case of methods 2, 3 and 4), such risks manifest themselves as rare events. In Figure C.1, the vertical threshold of 3 log10 cycles was placed on the graph for comparison with the target reduction.
For method 7, the mildness of the heat regimes of scenario 1 is evident in the respective lethality cumulative distributions of Figure C.2. This graph shows that the lethality against parvoviruses is higher in scenario 3, followed by scenario 2, scenario 4 and scenario 5, with scenario 1 on the left side of the threshold.
Appendix D – Report on expert knowledge elicitation
Description and methodology
The EKE (expert knowledge elicitation) questions concerned one hazard (C. perfringens spores) and nine methods (processes), making a total of nine data points to be assessed (Table D.1).
Table D.1.
Processes assess in the EKE
| Data point | Process | t/T combinations |
|---|---|---|
| 1 | Method 2 – coincidental | 100°C × 5′ – 110°C × 70′ – 120°C × 50′ |
| 2 | Method 3 – coincidental | 100°C × 40′ – 110°C × 42′– 120°C × 13′ |
| 3 | Method 4 – coincidental | 100°C × 3′ – 110°C × 5′ – 120°C × 5′ – 130°C × 3′ |
| 4 | Method 5 – coincidental | 80°C × 60′ – 100°C × 60′ |
| 5 | Method 7A – scenario 1 | 80°C × 14′ |
| 6 | Method 7B – scenario 2 | 95°C × 90′ |
| 7 | Method 7C – scenario 3 | 115°C × 56′ |
| 8 | Method 7D – scenario 4 | 125°C × 10′ |
| 9 | Method 7E – scenario 5 | 133°C × 5′ |
The EKE question was specified as follows: ‘What is the probability that a 5 log 10 reduction of spores of Clostridium perfringens is achieved, in more than 99% of cases, by application of each of the relevant processes (methods 2, 3, 4, 5 in coincidental mode and five t/T combinations of method 7), assuming that the processes are performed as prescribed and that the indicated process conditions are achieved?’
It is assumed that the standard process is correctly performed, under the conditions indicated by the process parameters, and as described in the opinion. Variability in process performance is not to be considered in this assessment. However, even without any variation in process performance, the log10 reduction achieved will vary to some extent from case to case. The question to answer is whether the target log10 reduction will be achieved in more than 99% of cases, because 100% may be too unrealistic and would become dependent on ‘exceptional cases’. Thus, the ‘probability’ in the question refers to uncertainty, not variability. Specifically, it expresses the degree of certainty that the target log10 reduction will be achieved in more than 99% of the cases.
The EKE consisted of two steps:
Step 1: individual judgements (4 April to 13 April 2023)
Step 2: consensus judgement (19 April 2023)
The experts comprised six Working Group (WG) members developing the opinion, plus one EFSA scientist who was supporting the WG. The elicitation was facilitated by an elicitor (hearing expert). A member of the EFSA scientific staff was appointed as rapporteur.
The EKE section was recorded, only as a support to prepare the notes. This recording has been deleted to assure anonymity of the experts.
Step 1: Individual judgements
Training was delivered to all participants on the general concept of probability, EFSA's approximate probability scale, uncertainty, variability and EKE.
During Step 1, the participants had 1 week to provide individual judgements for each of the nine processes, taking into account the version of the draft opinion at the beginning of the process (4 April 2023) with the evidence on thermal inactivation of the C. perfringens spores (including the data and modelling results), the description of the processes and the integration of the evidence and the uncertainty table, as well as the personal expertise and assessment of the uncertainties involved. To perform the individual judgements, the experts received by e‐mail a spreadsheet with a template to provide their answers. They did not discuss their judgements with other experts at this stage.
The answer for each process was given as a probability range that reflects the expert's degree of certainty that the indicated log10 reduction is achieved. These probability ranges could be one of those given in the approximate probability scale presented in EFSA's uncertainty guidance (Table D.2) or any other. The participants were encouraged to give explanations of the reasons for each subjective probability range.
Table D.2.
Probability scores proposed to the experts
| 99–100% (almost certain) |
| 95–99% (extremely likely) |
| 90–95% (very likely) |
| 66–90% (likely) |
| 33–66% (about as likely as not) |
| 10–33% (unlikely) |
| 5–10% (very unlikely) |
| 1–5% (extremely unlikely) |
| 0–1% (almost impossible) |
| 100% (certain) |
| 50–100% (more likely than not) |
| 0–50% (more unlikely than likely) |
| 0–100% (inconclusive) |
| Other (to be defined by the participant) |
The options included in the template were:
Step 2: Consensus judgement
The next step was to reach a consensus judgement for the datapoints of each of the nine processes during the open session in the WG meeting. It was explained that the consensus is not an average of the individual judgements, or a compromise where some experts defer to the judgement of other participants. The experts were asked to consider what a rational impartial observer (RIO) would judge, having considered the evidence, uncertainties, the individual judgements and having heard the discussion. Consensus can be any probability range, not necessarily one from the table with standard subjective probability ranges.
After getting an overview of the individual expert judgements obtained in step 1, at the beginning of the EKE session, the participants expressed the rationale behind their individual judgements, to clarify potential generic biases in their judgements.
The stepwise approach applied for each of the combinations consisted of the following actions:
Recall the processes and time/temperature combinations and target log10 reduction (i.e. 5 log10).
Display graphs showing expert ranges.
Invite some experts (most deviating) to explain the reasoning for their judgements.
Invite experts to review/revise own judgements.
Propose a consensus judgement and ask experts if this range reflects what a RIO would think. If needed, discuss and agree on the upper of lower range of the consensus judgement.
Check that the notes taken have captured the key reasons for the consensus judgements. responses to the proposed consensus ranges for the indicators
Results of Step 1: Individual judgements
Individual judgements were obtained from seven experts. The results are illustrated in Figures D.1 and D.2. Both figures show that there is large agreement between individual experts for some of the processes, and large disagreement for others. These results were discussed during Step 2 of the EKE.
Figure D.1.
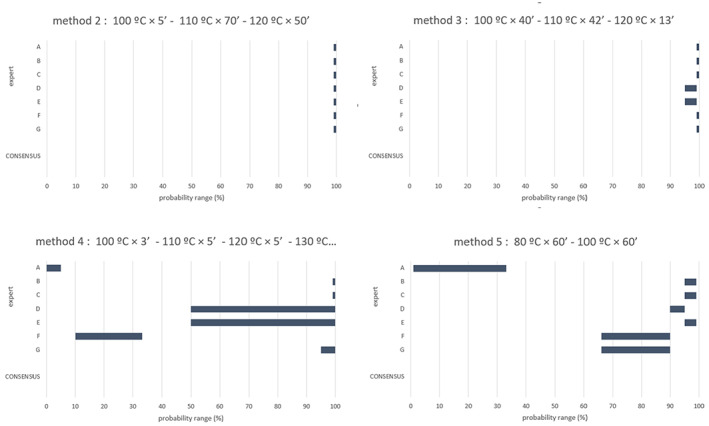
The individual elicited probability ranges by each of the seven experts for achieving a probability of 5 log10 reduction of spores of Clostridium perfringens, in more than 99% of cases, by application of methods 2, 3, 4, 5 in coincidental mode, assuming that the processes are performed as prescribed and that the indicated process conditions are achieved
Figure D.2.
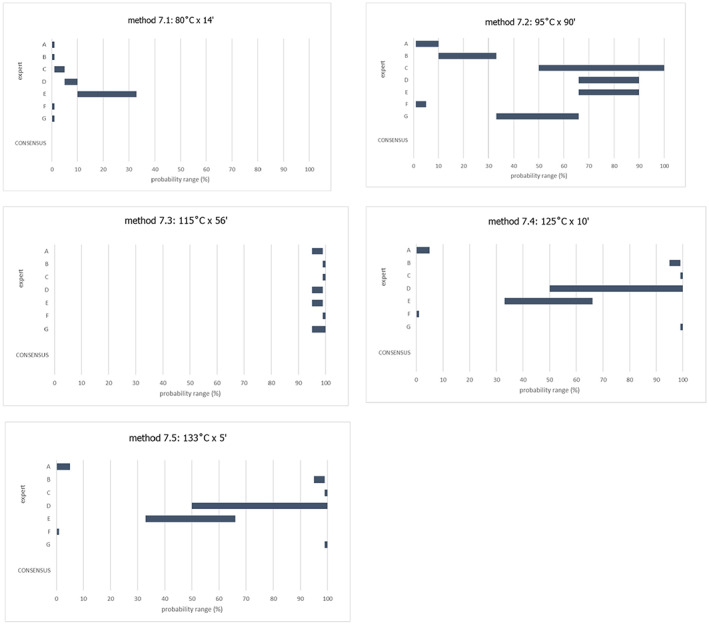
The individual elicited probability ranges by each of the seven experts for achieving a probability of 5 log10 reduction of spores of Clostridium perfringens, in more than 99% of cases, by application of the five t/T combinations of method 7, assuming that the processes are performed as prescribed and that the indicated process conditions are achieved
Individual expert's rationales for the individual judgements, as discussed at the start of Step 2
Expert A based the judgements mainly on the output of the model with and without temperature extrapolation, was very strict, and considered the judgements made as too conservative. Inactivation of spores of C. botulinum was also considered.
Expert B used the model outputs with and without extrapolation. This expert handled the data and has run the model with subsets of data and considers that the judgements therefore may be biased.
Expert C based the judgement on the model output without extrapolation as primary source considering there could be underestimation. The expert also considered inactivation of spores of C. botulinum when the process used high temperatures and short times as in these cases the model output could be far from reality. The expert explained that spores of C. botulinum could be used as a surrogate for spores of C. perfringens as these are more heat resistant. The expert considered that a 12 log10 reduction of spores of C. botulinum is accepted by public health authorities (equivalent to 3 min at 121.1°C). If that is accomplished, the expert considered that at least a 5 log10 reduction of spores of C. perfringens is achieved.
Expert D considered the output of the model in relation to the achievement of at least 5 log10 and between 5 and 6 log10 reduction of spores of C. perfringens.
Expert E based the judgement on the output of the model without extrapolation and considered that the model underestimates the inactivation. The expert did not consider inactivation of spores of C. botulinum.
Expert F based on judgement mainly on the output of the model.
Expert G used the entire information from the scientific opinion, not only the output of the model with and without extrapolation, but also the distribution of the D‐values at the relevant temperatures. The expert considered that the output of the model without extrapolation is underestimating the inactivation, depending on the method at hand (if the temperature is further from Dref, there is more underestimation).
Results of the consensus for each combination
After discussion among the experts, consensus was achieved on the probability ranges that were considered to best represent the uncertainty on whether 5 log10 reduction of spores of C. perfringens is achieved with each of the relevant processes. Consensus implied that the experts agreed that a RIO, considering the evidence and following the discussion, would conclude that the elicited probability range was appropriate.
Figure D.3 gives the outcome of the consensus reached for each process, while Table D.3 provides the main arguments for obtaining these ranges, for each of the nine processes.
Figure D.3.
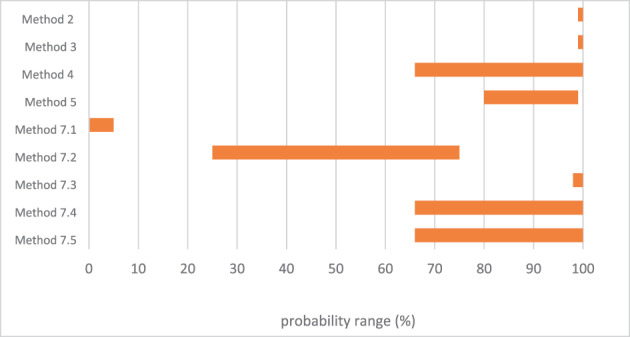
The consensus judgement for achieving a probability of 5 log10 reduction of spores of Clostridium perfringens, in more than 99% of cases, by application of the t/T combinations of methods 2–5 and method 7, assuming that the processes are performed as prescribed and that the indicated process conditions are achieved
Table D.3.
Summary of the rationale for the consensus of each process, as recorded by the rapporteur
| Process | Summary of rationale as recorded by the rapporteur |
|---|---|
|
Method 2 – coincidental 100°C × 5′ – 110°C × 70′ – 120°C × 50′ |
There was a full agreement between individual experts for this process; all experts provided individual elicited probability ranges of 99–100%. Expert C explained that the model output, without extrapolation, predicts more than 10 log10 inactivation of spores of C. perfringens, based on the time/temperature profile of the treatment. A higher inactivation is expected as the actual process temperature would be higher. The elicitator proposed a consensus judgement of 99–100% and this was agreed. |
|
Method 3 – coincidental 100°C × 40′ – 110°C × 42′ – 120°C × 13′ |
There was good agreement between individual experts for this process. Five experts provided individual elicited probability ranges of 99–100%, while two experts (expert D and E) provided ranges of 95–99%. Expert D gave a lower probability compared to method 2 considering that the processing time at 120°C is lower compared to the previous process. Expert C stated that the inactivation of spores of C. perfringens in method 3 would still be very high. The predicted inactivation, based on the model output, is beyond 6–8 log10 inactivation of spores of C. perfringens and the actual processing temperature would be higher than that. Expert G confirmed that the probability was 100% for at least a 5 log10 inactivation of spores of C. perfringens and as the temperature is higher than the temperature used for Dref (i.e. 105°C), the inactivation is underestimated. Experts D and E, after hearing the arguments of the other experts, agreed to increase their probability range to 99–100%. The elicitator proposed a consensus judgement of 99–100% and this was agreed. |
|
Method 4 – coincidental 100°C × 3′ – 110°C × 5′ – 120°C × 5′ – 130°C × 3′ |
There was a large disagreement between individual experts for this process. Expert A (providing a probability range of 0–5%) considered the high probability (82.1%) in the model output that a 5 log10 inactivation of spores of C. perfringens would not be achieved. The expert considered extrapolation. The expert informed that the probability could be revised to values higher than 50% if also the inactivation of spores of C. botulinum would be considered. Expert G (providing a probability range of 95–100%), stated that the probability was about 18% for at least a 5 log10 inactivation of C. perfringens, based on model predictions. There is a very high underestimation as high temperatures were used for short times. Even if C. perfringens is the focus, spores of C. botulinum are more resistant. Therefore, the expert is quite sure that if 5 log10 inactivation of spores of C. botulinum would be met, it would also be the case for spores of C. perfringens. Therefore, this expert increased the probability range, but judged a slightly lower probability range compared to method 3 considering the time/temperature (t/T) combinations. Expert B (providing a probability range of 99–100%) considered the model output using extrapolation as high temperatures were used in this process and the model without extrapolation is ‘truncated’ at 105°C (Tref). As the model did not consider the temperature come‐up time, the expert was certain that a 5 log10 inactivation of spores of C. perfringens is achieved. Expert C (providing a probability range of 99–100%) based the judgement on C. botulinum. The expert informed the group that the actual temperature is almost 30°C higher than Tref. An inactivation equivalent to more than 27 min at 121°C would be expected for spores of C. botulinum, sufficient to ensure 12 log10 reductions of those spores (3 min is enough). Therefore, the expert considered that inactivation of spores of C. perfringens would be almost certain. However, the expert proposes to broaden the probability range to increase the uncertainty. Expert F (providing a probability range of 10–33%) only considered C. perfringens and found it unlikely that there would be a 5 log10 inactivation of spores of C. perfringens, based on the model outputs. Expert A (providing a probability range of 0–5%) asked to agree on the extent to use the data on inactivation of spores of C. botulinum in the judgements. If so, it would become very likely to achieve a 5 log10 inactivation of spores of C. perfringens. Expert B informed that the probability range could be lowered from 99–100% to 95–100%. Expert E stated that the reasoning to consider C. botulinum as a surrogate/indicator for C. perfringens needs to be well explained in the opinion. Also, the underestimation of the model should be better explained in the opinion. This was agreed by expert G. Expert D provided a large probability range (of 50–100%) as the probability of at least a 5 log10 inactivation of spores of C. perfringens is low (about 18%), but there is high probability to achieve between 5 and 6 log10 inactivation of spores of C. perfringens. The expert would stay on the right side of the probability scale but use a range wider than 95–100%. Expert B was asked to explain the model outputs. The expert recalled that the model was truncated at 105°C. When temperatures are above 105°C, there is more uncertainty and underestimation (but not clear to what extent). Expert E then proposed to have a large probability range (of 50–100%), which was agreed by expert F. The elicitator proposed 66% as lower limit of the probability range which was agreed. The upper limit for some experts was 100% and it was agreed to use this value as an upper limit considering also C. botulinum spores' inactivation. Thus, the agreed consensus judgement was 66–100%. |
|
Method 5 – coincidental 80°C × 60′ – 100°C × 60′ |
There was again a large disagreement between individual experts for this process. Expert A (providing a probability range of 1–33%) based on discussions for other process, believes that spores of C. botulinum have low mortality here, so the expert is not positive to reach a 5 log10 inactivation of spores of C. perfringens in 99% of cases. Expert C (providing a probability range of 95–99%) explained that the predicted inactivation of the temperature profile is higher than 5 log10 with a 99% probability. The actual temperature is within the limits where there are published data available, so the prediction is expected to be close to reality. The inactivation is more likely than not but would broaden the range provided. Expert B (providing a probability range of 95–99%) said that the models output can be trusted without extrapolation considering the temperatures used in this process. Considering additionally the come‐up time, the provided range is believed still valid. This was agreed by expert E. Expert G (providing a probability range of 66–90%) believes the model overestimates the inactivation of spores of C. botulinum because the product would be quite dehydrated through the treatment. Expert F (providing a probability range of 66–90%) considered mainly the model outputs and expects explanations to be added in opinion. Expert A doubted about the upper range of 99% considering the model outputs. The elicitator explained that the model percentages are to be considered also uncertainties. The expert then agreed that ‘more likely than not’ should be the outcome. Adding uncertainty, the expert would agree with a probability range of 66–90%. The elicitator proposed 66% as lower limit of the probability range. According to expert B, it should be rather 90% as explained before, also considering dehydration of the product. Expert D agrees with 90% as lower limit considering the model output and the estimated inactivation of spores of C. perfringens between 5 and 6 log10. The agreed consensus judgement was 80–99%. |
|
Method 7 – scenario 1 80°C × 14′ |
There was some level of agreement between individual expert judgements for this process. Most extreme were four experts providing individual elicited probability ranges of 0–1%, while one expert (expert E) provided a range of 10–33%. Expert E wanted to give a lower probability as well but then considered the underestimation of the model (through the come‐up time). The expert would agree to lower the probability. Expert D explained that the probability was 100% for less than a 5 log10 inactivation of spores of C. perfringens, and the probability is 0% when between 5 to 6 log10. Expert C considers that method is quite insufficient (based on the t/T profiles) to inactivate spores of C. perfringens, but it also needs to be considered that the data used in the model relies on highly resistant spores. Only if all the spore population would have low heat resistance, a 5 log10 reduction could be accomplished. The agreed consensus judgement was 0–5%. |
|
Method 7 – scenario 2 95°C × 90′ |
There was again a large disagreement between individual experts for this process, with experts A and F providing the lowest probability ranges. The model output yields a probability of 31.9% that a 5 log10 inactivation of spores of C. perfringens would not be achieved, based on the t/T profile of the method. Expert F would agree to increase the probability range. Expert A informed that there is no need to extrapolate, and C. botulinum inactivation is not to be used. The probability would be rather below 50%. Expert E (providing a probability range of 66–90%) would rather have a probability beyond 50% considering the under‐estimation of the inactivation by the model. Expert C (providing a probability range of 50–100%) considers the probability more likely than not also considering there was no extrapolation needed. The expert would lower the upper limit to 90%. The elicitator questioned if the lower limit should be below or above 50%. According to expert B (providing a probability range of 10–33%), the range should be quite low (below 50%) as the mean inactivation of spores of C. perfringens is 7 log10. Expert G (providing a probability range of 33–66%), also considered that in most of the studies it needs more than 90′ to achieve a 5D inactivation of spores of C. perfringens. The elicitator then asked about upper limit. Expert C agreed to lower it to 90%. Expert B proposes 66%. Expert F proposed to use 33–66% as probability range as this would reflect that there is a high uncertainty. The elicitator questioned whether the upper limit is acceptable considering the model output (68.1% that a 5 log10 inactivation of spores of C. perfringens would be achieved); hence the model must be overestimating. Expert E further questioned the divergence with method 5 where we were more certain. According to expert C the impact of a small change in temperature is significant. A higher value of the upper range (80%) was proposed to reflect uncertainty. Finally expert C proposed 25–75%, which was agreed as consensus judgement. |
|
Method 7 – scenario 3 115°C × 56′ |
There was a high level of agreement between individual experts for this process; all experts provided high probability ranges. The model output yields a probability of 100% that a 5 log10 inactivation of spores of C. perfringens would be achieved, based on the t/T profile of the method. Expert C (providing a probability range of 99–100%) had a high level of confidence as the predicted inactivation based on the t/T profile is higher than 6–8 log10 and considered that truncation was applicable as the actual temperature is higher. The method would reach 12 log10 reduction of spores of C. botulinum, with an equivalent time at 121°C of 13.7 min. Expert F (providing a probability range of 99–100%) followed the same reasoning based on the t/T profile. Expert E (providing a probability range of 95–99%) considered that the t/T profile is a bit less stringent than method 2 and therefore gave somewhat lower values. The expert informed that the probability could be increased. Expert G (providing a probability range of 95–100%) said that 100% should be included in the range. Expert A (providing a probability range of 95–99%) had the same arguments. The upper limit of 100% as consensus judgement was agreed by all. There was no clear argument for underestimation of the model and 98% as lower limit was agreed. |
|
Method 7 – scenario 4 125°C × 10′ |
There was a large disagreement between individual experts for this process, with experts A and F again providing the lowest probability ranges. The model output yields a probability of 98.5% that a 5 log10 inactivation of spores of C. perfringens would not be achieved, based on the t/T profile of the method. In this process higher temperatures are used, so there is an extrapolation effect. Expert G (providing a probability range of 99–100%), as in method 4, indicated that there is a huge underestimation in the model outputs as the process relies on high temperature/short time processing and considered inactivation of spores of C. botulinum. The expert would consider revising the probability range to the consensus reached for method 4. Expert C (providing a probability range of 99–100%) followed the same reasoning. When inactivation of spores of C. botulinum is achieved an inactivation equivalent to more than 24.7 min at 121°C would be expected, sufficient to ensure 12 log reductions of C. botulinum, the expert gave a high probability, similarly to method 4. Expert A (providing a probability range of 0–5%) agreed and would be fine to revise the range as agreed for method 4. Expert F (providing a probability range of 0–1%) agreed as well. Expert E (providing a probability range of 33–66%) is in middle of the probability range. The expert is uncertain, and the arguments are the same. The model predictions are high but there is a larger underestimation. The expert did not consider C. botulinum, but only the model output and underestimation. The elicitator asked if a comparable range to method 4, method 7 scenario 4 and method 7 scenario 5 would apply (66–100%) as the arguments are the same. Expert B (providing a probability range of 95–99%) would not include 100% as the highest temperature is different and there are more come‐up times in method 4. Expert C believes that 100% is reasonable as these conditions would lead to a safe process (accepted as a safe for pet food in legislation). The elicitator proposed a consensus judgement of 66–100% and this was agreed |
|
Method 7 – scenario 5 133°C × 5′ |
The model output yields a probability of 100% that a 5 log10 inactivation of spores of C. perfringens would not be achieved, based on the t/T profile of the method. The elicitator proposed for this process the same consensus judgement as for method 7 scenario 5 (66–100%) comparing the t/T profiles of both methods and the individual expert judgements. This was agreed. |
Annex A – Protocol for the assessment of the efficacy of methods 2 to 5 and method 7 to inactivate relevant pathogens when producing processed animal protein of porcine origin intended to feed poultry and aquaculture animals (EFSA‐Q‐2022‐00455)
1.
Protocol for the assessment of the efficacy of methods 2 to 5 and method 7 to inactivate relevant pathogens when producing processed animal protein of porcine origin intended to feed poultry and aquaculture animals (EFSA‐Q‐2022‐00455) is available under the Supporting Information section on the online version of the scientific output.
Supporting information
Protocol for the assessment of the efficacy of methods 2 to 5 and method 7 to inactivate relevant pathogens when producing processed animal protein of porcine origin intended to feed poultry and aquaculture animals (EFSA‐Q‐2022‐00455)
Suggested citation: EFSA BIOHAZ Panel (EFSA Panel on Biological Hazards) , Koutsoumanis K, Allende A, Alvarez Ordoñez A, Bolton D, Bover‐Cid S, Chemaly M, Herman L, Hilbert F, Lindqvist R, Nauta M, Nonno R, Peixe L, Skandamis P, Suffredini E, Fernandez Escamez P, Gonzales‐Barron U, Roberts H, Ru G, Simmons M, Barcia Cruz R, Lourenço Martins J, Messens W, Ortiz‐Pelaez A, Simon AC and De Cesare A, 2023. Scientific Opinion on the assessment on the efficacy of methods 2 to 5 and method 7 set out in Commission Regulation (EU) No 142/2011 to inactivate relevant pathogens when producing processed animal protein of porcine origin intended to feed poultry and aquaculture animals. EFSA Journal 2023;21(7):8093, 76 pp. 10.2903/j.efsa.2023.8093
Requestor European Commission
Question number EFSA‐Q‐2022‐00455
Panel members Ana Allende, Avelino Alvarez‐Ordoñez, Declan Bolton, Sara Bover‐Cid, Marianne Chemaly, Alessandra De Cesare, Lieve Herman, Friederike Hilbert, Konstantinos Koutsoumanis, Roland Lindqvist, Maarten Nauta, Romolo Nonno, Luisa Peixe, Giuseppe Ru, Marion Simmons, Panagiotis Skandamis and Elisabetta Suffredini.
Declarations of interest If you wish to access the declaration of interests of any expert contributing to an EFSA scientific assessment, please contact interestmanagement@efsa.europa.eu.
Acknowledgements The BIOHAZ Panel wishes to acknowledge the European Fat processors and Renderers Association (EFPRA) for providing data, and Kateryna Chuzhakina for her contribution to the production of this opinion.
EFSA may include images or other content for which it does not hold copyright. In such cases, EFSA indicates the copyright holder and users should seek permission to reproduce the content from the original source.
Adopted: 7 June 2023
Notes
Commission Regulation (EU) 2021/1372 of 17 August 2021 amending Annex IV to Regulation (EC) No 999/2001 of the European Parliament and of the Council as regards the prohibition to feed non‐ruminant farmed animals, other than fur animals, with protein derived from animals. OJ L 295, 18.8.2021, p. 1.
Commission Regulation (EU) No 142/2011 of 25 February 2011 implementing Regulation (EC) No 1069/2009 of the European Parliament and of the Council laying down health rules as regards animal by‐products and derived products not intended for human consumption and implementing Council Directive 97/78/EC as regards certain samples and items exempt from veterinary checks at the border under that Directive. OJ L 54, 26.2.2011, p. 1.
Regulation (EU) 2019/1009 of the European Parliament and of the Council of 5 June 2019 laying down rules on the making available on the market of EU fertilising products and amending Regulations (EC) No 1069/2009 and (EC) No 1107/2009 and repealing Regulation (EC) No 2003/2003. OJ L 170, 25.6.2019, p. 1–114.
Regulation (EC) No 178/2002 of the European Parliament and of the Council of 28 January 2002 laying down the general principles and requirements of food law, establishing the European Food Safety Authority and laying down procedures in matters of food safety. OJ L 31, 1.2.2002, p. 1.
According to Commission Regulation (EU) 142/2011, Annex XIII, Chapter II, point 3 ‘canned petfood must be subjected to heat treatment to a minimum Fc value of 3’.
ECDC, online. Trichinellosis. Available online: https://www.ecdc.europa.eu/en/trichinellosis
Commission Regulation (EC) No 2075/2005 of 5 December 2005 laying down specific rules on official controls for Trichinella in meat (Text with EEA relevance). OJ L 338, 22.12.2005, p. 60., as amended.
References
- Abworo EO, Onzere C, Amimo JO, Riitho V, Mwangi W, Davies J, Blome S and Bishop RP, 2017. Detection of African swine fever virus in the tissues of asymptomatic pigs in smallholder farming systems along the Kenya‐Uganda border: implications for transmission in endemic areas and ASF surveillance in East Africa. Journal of General Virology, 98, 1806–1814. [DOI] [PubMed] [Google Scholar]
- Adedeji AJ, Atai RB, Gyang HE, Gambo P, Habib MA, Weka R, Muwanika VB, Masembe C and Luka PD, 2022. Live pig markets are hotspots for spread of African swine fever virus in Nigeria. Transboundary and Emerging Diseases, 69, e1526–e1540. [DOI] [PubMed] [Google Scholar]
- Aguirre J, Pin C, Rodriguez M and Garcia de Fernando G, 2009. Analysis of the variability in the number of viable bacteria after mild heat treatment of food. Applied and Environmental Microbiology, 75, 6992–6997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Almeida MN, Zimmerman JJ, Wang C and Linhares DCL, 2018. Assessment of abattoir based monitoring of PRRSV using oral fluids. Preventive Veterinary Medicine, 158, 137–145. [DOI] [PubMed] [Google Scholar]
- Amorim AR, Fornells L, Reis FD, Rezende DJ, Mendes GD, Couceiro J and Santos NSD, 2013. Influenza A virus infection of healthy piglets in an abattoir in Brazil: animal‐human interface and risk for interspecies transmission. Memorias Do Instituto Oswaldo Cruz, 108, 548–553. 10.1590/0074-0276108052013003 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Amorim AR, Mendes GS, Pena GPA and Santos N, 2018. Hepatitis E virus infection of slaughtered healthy pigs in Brazil. Zoonoses and Public Health, 65, 501–504. 10.1111/zph.12455 [DOI] [PubMed] [Google Scholar]
- Andersen KG, Hansen TB and Knochel S, 2004. Growth of heat‐treated enterotoxin‐positive Clostridium perfringens and the implications for safe cooling rates. Journal of Food Protection, 67, 83–89. 10.4315/0362-028x-67.1.83 [DOI] [PubMed] [Google Scholar]
- Aryal M and Liu G, 2021. Porcine Bocavirus: a 10‐year history since its discovery. Virologica Sinica, 36, 1261–1272. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Baudon E, Poon LL, Dao TD, Pham NT, Cowling BJ, Peyre M, Nguyen KV and Peiris M, 2015. Detection of Novel Reassortant Influenza A (H3N2) and H1N1 2009 Pandemic Viruses in Swine in Hanoi. Vietnam. Zoonoses and Public Health, 62, 429–434. 10.1111/zph.12164 [DOI] [PubMed] [Google Scholar]
- Baudon E, Chu DKW, Dao Duy T, Pham Thi N, Mai P HV, Nguyen Le Khanh H, Le Thi T, Nguyen Thanh T, Nguyen Cong K, LeQuynh M, Nguyen Viet K, Cowling BJ, Peyre M and Peiris M, 2018. Swine influenza viruses in Northern Vietnam in 2013–2014. Emerging Microbes and Infections, 7, 123. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Benfield DA and Hesse RA, 2019. Adenoviruses. Diseases of Swine. 438–442 [Google Scholar]
- Benkő M, Aoki K, Arnberg N, Davison AJ, Echavarría M, Hess M, Jones MS, Kaján GL, Kajon AE and Mittal SK, 2022. ICTV virus taxonomy profile: Adenoviridae 2022. Journal of General Virology, 103, 001721. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Berto A, Martelli F, Grierson S and Banks M, 2012. Hepatitis E Virus in Pork Food Chain, United Kingdom, 2009–2010. Emerging Infectious Diseases, 18, 1358–1360. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bigelow W, 1921. The logarithmic nature of thermal death time curves. The Journal of Infectious Diseases, 29, 528–536. [Google Scholar]
- Bigoraj E, Paszkiewicz W and Rzezutka A, 2021. Porcine blood and liver as sporadic sources of Hepatitis E Virus (HEV) in the production chain of offal‐derived foodstuffs in Poland. Food and Environmental Virology, 13, 347–356. 10.1007/s12560-021-09475-z [DOI] [PMC free article] [PubMed] [Google Scholar]
- Blazquez E, Rodriguez C, Rodenas J, Navarro N, Rosell R, Pina‐Pedrero S, Campbell JM, Sibila M, Segales J, Pujols J and Polo J, 2019. UV‐C irradiation is able to inactivate pathogens found in commercially collected porcine plasma as demonstrated by swine bioassay. Veterinary Microbiology, 239, 5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Blázquez E, Pujols J, Segalés J, Rodríguez C, Campbell J, Russell L and Polo J, 2022. Estimated quantity of swine virus genomes based on quantitative PCR analysis in spray‐dried porcine plasma samples collected from multiple manufacturing plants. Plos One, 17, e0259613. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Blomström A‐L, Belák S, Fossum C, Fuxler L, Wallgren P and Berg M, 2010. Studies of porcine circovirus type 2, porcine boca‐like virus and torque teno virus indicate the presence of multiple viral infections in postweaning multisystemic wasting syndrome pigs. Virus research, 152, 59–64. [DOI] [PubMed] [Google Scholar]
- Blumel J, Schmidt I, Effenberger W, Seitz H, Willkommen H, Brackmann HH, Lower J and Eis‐Hubinger AM, 2002. Parvovirus B19 transmission by heat‐treated clotting factor concentrates. Transfusion, 42, 1473–1481. 10.1046/j.1537-2995.2002.00221.x [DOI] [PubMed] [Google Scholar]
- Boniotti MB, Papetti A, Bertasio C, Giacomini E, Lazzaro M, Cerioli M, Faccini S, Bonilauri P, Vezzoli F, Lavazza A and Alborali GL, 2018. Porcine epidemic diarrhoea virus in Italy: disease spread and the role of transportation. Transboundary and Emerging Diseases, 65, 1935–1942. 10.1111/tbed.12974 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bouquet J, Tesse S, Lunazzi A, Eloit M, Rose N, Nicand E and Pavio N, 2011. Close similarity between sequences of Hepatitis E virus recovered from humans and Swine, France, 2008–2009. Emerging Infectious Diseases, 17, 2018–2025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Boxman ILA, Jansen CCC, Hagele G, Zwartkruis‐Nahuis A, Cremer J, Vennema H and Tijsma ASL, 2017. Porcine blood used as ingredient in meat productions may serve as a vehicle for hepatitis E virus transmission. International Journal of Food Microbiology, 257, 225–231. [DOI] [PubMed] [Google Scholar]
- Boxman ILA, Verhoef L, Dop PY, Vennema H, Dirks RAM and Opsteegh M, 2022. High prevalence of acute hepatitis E virus infection in pigs in Dutch slaughterhouses. International Journal of Food Microbiology, 379, 10. [DOI] [PubMed] [Google Scholar]
- Brauniger S, Peters J, Borchers U and Kao M, 2000. Further studies on thermal resistance of bovine parvovirus against moist and dry heat. International Journal of Hygiene and Environmental Health, 203, 71–75. 10.1078/s1438-4639(04)70010-3 [DOI] [PubMed] [Google Scholar]
- Brooks A, 2013. Wet‐heat inactivation of bacterial endospers in packaged fruit juices. International Journal of Current Microbiology and Applied Sciences, 2, 506–515. [Google Scholar]
- Burtscher C and Wuertz S, 2003. Evaluation of the use of PCR and reverse transcriptase PCR for detection of pathogenic bacteria in biosolids from anaerobic digestors and aerobic composters. Applied and Environmental Microbiology, 69, 4618–4627. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Byrne B, Dunne G and Bolton DJ, 2006. Thermal inactivation of Bacillus cereus and Clostridium perfringens vegetative cells and spores in pork luncheon roll. Food Microbiology, 23, 803–808. 10.1016/j.fm.2006.02.002 [DOI] [PubMed] [Google Scholar]
- Cadar D, Cságola A, Kiss T and Tuboly T, 2013. Capsid protein evolution and comparative phylogeny of novel porcine parvoviruses. Molecular Phylogenetics and Evolution, 66, 243–253. [DOI] [PubMed] [Google Scholar]
- Cappai MG, Rubiu NG and Pinna W, 2018. Economic assessment of a smart traceability system (RFID plus DNA) for origin and brand protection of the pork product labelled “suinetto di Sardegna”. Computers and Electronics in Agriculture, 145, 248–252. [Google Scholar]
- Carabin H and Traoré AA, 2014. Taenia solium taeniasis and cysticercosis control and elimination through community‐based interventions. Current Tropical Medicine Reports, 1, 181–193. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Casas M, Cortes R, Pina S, Peralta B, Allepuz A, Cortey M, Casal J and Martin M, 2011. Longitudinal study of hepatitis E virus infection in Spanish farrow‐to‐finish swine herds. Veterinary Microbiology, 148, 27–34. [DOI] [PubMed] [Google Scholar]
- Chambaro HM, Sasaki M, Muleya W, Kajihara M, Shawa M, Mwape KE, Harima H, Qiu YJ, Hall WW, Fandamu P, Squarre D, Simulundu E, Sawa H and Orba Y, 2021. Hepatitis E virus infection in pigs: a first report from Zambia. Emerging Microbes and Infections, 10, 2169–2172. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chelli E, Suffredini E, De Santis P, De Medici D, Di Bella S, D'Amato S, Gucciardi F, Guercio A, Ostanello F, Perrone V, Purpari G, Scavia GS, Schembri P, Varcasia BM and Di Bartolo I, 2021. Hepatitis E virus occurrence in pigs slaughtered in Italy. Animals, 11, 14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen R, Lai T, Chen Q, Wu X, Che Y, Yan S, Wang C, Wang L and Zhou L, 2018. Genetic characterization of porcine parvovirus 7 (PPV7) isolates in Fujian, China. Kafkas Universitesi Veteriner Fakultesi Dergisi, 24, 473–477. [Google Scholar]
- Chen LB, Wen K, Chen FE, Trick AY, Liu HB, Shao SB, Yu WB, Hsieh KW, Wang ZH, Shen JZ and Wang TH, 2021. Portable magnetofluidic device for point‐of‐need detection of African swine fever. Analytical Chemistry, 93, 10940–10946. [DOI] [PubMed] [Google Scholar]
- Cheung JTL, Lau EHY, Jin ZY, Zhu HC, Guan Y and Peiris M, 2022. Influenza A virus transmission in swine farms and during transport in the swine supply chain. Transboundary and Emerging Diseases, 69, E3101–E3110. 10.1111/tbed.14667 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cho KH, Kim HJ, Jang MK, Ryu JH, Yoo D, Kang HE and Park JY, 2022. Detection of African Swine Fever at an Abattoir in South Korea, 2020. Veterinary Sciences, 9, 7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cortey M, Macera L, Segalés J and Kekarainen T, 2011. Genetic variability and phylogeny of Torque teno sus virus 1 (TTSuV1) and 2 (TTSuV2) based on complete genomes. Veterinary Microbiology, 148, 125–131. [DOI] [PubMed] [Google Scholar]
- Craven SE, 1990. The effect of the pH of the sporulation environment on the heat resistance of Clostridium perfringens spores. Current Microbiology, 20, 233–237. [Google Scholar]
- Crisostomo‐Jorquera V and Landaeta‐Aqueveque C, 2022. The genus Trichinella and its presence in wildlife worldwide: a review. Transboundary and Emerging Diseases, 69, E1269–E1279. 10.1111/tbed.14554 [DOI] [PubMed] [Google Scholar]
- Csagola A, Kiss I and Tuboly T, 2008. Detection and analysis of porcine circovirus type 1 in Hungarian wild boars: short communication. Acta Veterinaria Hungarica, 56, 139–144. [DOI] [PubMed] [Google Scholar]
- Da Silva MS, Budaszewski RF, Weber MN, Cibulski SP, Paim WP, Mosena ACS, Canova R, Varela APM, Mayer FQ, Pereira CW and Canal CW, 2020. Liver virome of healthy pigs reveals diverse small ssDNA viral genomes. Infection Genetics and Evolution, 81, 13. [DOI] [PubMed] [Google Scholar]
- de Arruda LR, Ribeiro J, Alfieri AF and Alfieri AA, 2013. Simultaneous infection with distinct strains of torque teno sus virus (TTSuV) in healthy slaughter‐age pigs. Veterinary Research Communications, 37, 183–186. [DOI] [PubMed] [Google Scholar]
- De Conti ER, Takeuti KL, Schwertz CI, Bianchi RM, Driemeier D and e Barcellos D, 2021. Agents of pneumonia in slaughtered pigs in southern Brazils. Pesquisa Veterinaria Brasileira, 41, 7. [Google Scholar]
- de Paula VS, Wiele M, Mbunkah AH, Daniel AM, Kingsley MT and Schmidt‐Chanasit J, 2013. Hepatitis E virus genotype 3 strains in domestic pigs, Cameroon. Emerging Infectious Diseases, 19, 686–688. [DOI] [PMC free article] [PubMed] [Google Scholar]
- De Souza AJS, Gomes‐Gouvea MS, Soares MDP, Pinho JRR, Malheiros AP, Carneiro LA, os Santos DRL and Pereira WLA, 2012. HEV infection in swine from Eastern Brazilian Amazon: evidence of co‐infection by different subtypes. Comparative Immunology Microbiology and Infectious Diseases, 35, 477–485. [DOI] [PubMed] [Google Scholar]
- Di Bartolo I, Diez‐Valcarce M, Vasickova P, Kralik P, Hernandez M, Angeloni G, Ostanello F, Bouwknegt M, Rodriguez‐Lazaro D, Pavlik I and Ruggeri FM, 2012. Hepatitis E virus in pork production chain in Czech Republic, Italy, and Spain, 2010. Emerging Infectious Diseases, 18, 1282–1289. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Di Martino B, Di Profio F, Martella V, Di Felice E, Di Francesco CE, Ceci C and Marsilio F, 2010. Detection of hepatitis E virus in slaughtered pigs in Italy. Archives of Virology, 155, 103–106. 10.1007/s00705-009-0544-0 [DOI] [PubMed] [Google Scholar]
- Diao MM, André S and Membré J‐M, 2014. Meta‐analysis of D‐values of proteolytic Clostridium botulinum and its surrogate strain Clostridium sporogenes PA 3679. International Journal of Food Microbiology, 174, 23–30. [DOI] [PubMed] [Google Scholar]
- dos Santos DRL, de Paula VS, de Oliveira JM, Marchevsky RS and Pinto MA, 2011. Hepatitis E virus in swine and effluent samples from slaughterhouses in Brazil. Veterinary Microbiology, 149, 236–241. 10.1016/j.vetmic.2010.10.024 [DOI] [PubMed] [Google Scholar]
- Doyle ME and Mazzotta AS, 2000. Review of studies on the thermal resistance of Salmonellae. Journal of Food Protection, 63, 779–795. [DOI] [PubMed] [Google Scholar]
- Ducatez MF, Awoume F and Webby RJ, 2015. Influenza A(H1N1)pdm09 virus in pigs, Togo, 2013. Veterinary Microbiology, 177, 201–205. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dzierzon J, Oswaldi V, Merle R, Langkabel N and Meemken D, 2022. Hepatitis E virus cross‐contamination on the surface of porcine livers after storage in Euro meat containers in a German pig abattoir. Journal of Consumer Protection and Food Safety, 17, 33–39. 10.1007/s00003-021-01357-7 [DOI] [Google Scholar]
- Ebwanga EJ, Ghogomu SM and Paeshuyse J, 2022. Molecular characterization of ASFV and differential diagnosis of Erysipelothrix in ASFV‐infected pigs in pig production regions in Cameroon. Veterinary Sciences, 9, 15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- EFSA (European Food Safety Authority) , 2005a. Opinion on the safety vis‐à‐vis biological risks of biogas and compost treatment standards of animal by‐products. EFSA Journal 2005:264, 21 pp. 10.2903/j.efsa.2005.264 [DOI] [Google Scholar]
- EFSA (European Food Safety Authority) , 2005b. Opinion of the Scientific Panel on Biological Hazards of the European Food Safety Authority on the biological safety of heat treatment of manure. Available online: 10.2903/j.efsa.2005.265 [DOI]
- EFSA AHAW Panel (EFSA Panel on Animal Health and Welfare) , Nielsen SS, Alvarez J, Bicout DJ, Calistri P, Canali E, Drewe JA, Garin‐Bastuji B, Gonzales Rojas JL, Schmidt CG, Herskin M, Michel V, Miranda Chueca MA, Padalino B, Pasquali P, Stahl K, Calvo AV, Viltrop A, Winckler C, De Clercq K, Sjunnesson Y, Gervelmeyer A and Roberts HC, 2022. Scientific Opinion on the assessment of the control measures of the Category A diseases of the Animal Health Law: prohibitions in restricted zones and risk‐mitigating treatments for products of animal origin and other materials. EFSA Journal 2022;20(8):7443, 128 pp. 10.2903/j.efsa.2022.7443 [DOI] [PMC free article] [PubMed] [Google Scholar]
- EFSA (European Food Safety Authority), Martino L, Aiassa E, Halldórsson TI, Koutsoumanis PK, Naegeli H, Baert K, Baldinelli F, Devos Y, Lodi F, Lostia A, Manini P, Merten C, Messens W, Rizzi V, Tarazona J, Titz A, Vos S, 2020. Draft framework for protocol development for EFSA’s scientific assessments. EFSA supporting publication 2020:EN‐1843. 46 pp. https://doi.org/10.2903/sp.efsa.2020.EN‐1843
- EFSA BIOHAZ Panel (EFSA Panel on Biological Hazards) , Koutsoumanis K, Allende A, Bolton D, Bover‐Cid S, Chemaly M, Davies R, De Cesare A, Herman L, Hilbert F, Lindqvist R, Nauta M, Peixe L, Ru G, Simmons M, Skandamis P, Suffredini E, Bottari B, Cummins E, Ylivainio K, Munoz Guajardo I, Ortiz‐Pelaez A and Alvarez‐Ordoñez A, 2021. Inactivation of indicator microorganisms and biological hazards by standard and/or alternative processing methods in Category 2 and 3 animal by‐products and derived products to be used as organic fertilisers and/or soil improvers. EFSA Journal 2021;19(12):6932, 111 pp. 10.2903/j.efsa.2021.6932 [DOI] [PMC free article] [PubMed] [Google Scholar]
- EFSA BIOHAZ Panel (EFSA Panel on Biological Hazards), Koutsoumanis K, Allende A, Bolton D, Bover–Cid S, Chemaly M, Davies R, De Cesare A, Herman L, Hilbert F, Lindqvist R, Nauta M, Peixe L, Ru G, Simmons M, Skandamis P, Suffredini E, Fernandez Escamez P, Griffin J, Ortiz Pelaez A and Alvarez–Ordonez A, 2022. Scientific Opinion on the evaluation of a multi–step catalytic coprocessing hydrotreatment for the production of renewable fuels using Category 3 animal fat and used cooking oils. EFSA Journal 2022;20(11):7591, 42 pp. https://doi.org/10.2903/j.efsa.2022.7591 [DOI] [PMC free article] [PubMed]
- Elving J, Vinnerås B, Albihn A and Ottoson JR, 2014. Thermal treatment for pathogen inactivation as a risk mitigation strategy for safe recycling of organic waste in agriculture. Journal of Environmental Science and Health, Part B, 49, 679–689. [DOI] [PubMed] [Google Scholar]
- Evelyn X and Silva FVM, 2015. Use of power ultrasound to enhance the thermal inactivation of Clostridium perfringens spores in beef slurry. International Journal of Food Microbiology, 206, 17–23. 10.1016/j.ijfoodmicro.2015.04.013 [DOI] [PubMed] [Google Scholar]
- Feng Z, Hirai‐Yuki A, McKnight KL and Lemon SM, 2014. Naked viruses that aren't always naked: quasi‐enveloped agents of acute hepatitis. Annual Review of Virology, 1, 539–560. [DOI] [PubMed] [Google Scholar]
- Feurer C, Le Roux A, Rossel R, Barnaud E, Dumarest M, Garry P and Pavio N, 2018. High load of hepatitis E viral RNA in pork livers but absence in pork muscle at French slaughterhouses. International Journal of Food Microbiology, 264, 25–30. [DOI] [PubMed] [Google Scholar]
- Florez J, Vera V, Lora A and Ramirez‐Nieto G, 2018. Molecular evaluation of influenza A virus in swine at slaughterhouses in Colombia. Revista Mvz Cordoba, 23, 7013–7024. [Google Scholar]
- Forero JE, Gutiérrez ‐Vergara C, Suescun JP, Correa G, Rodriguez B, Gutiérrez LA, Diaz FJ and Lopez‐Herrera A, 2017. Phylogenetic analysis of Hepatitis E virus strains isolated from slaughter‐age pigs in Colombia. Infection Genetics and Evolution, 49, 138–145. [DOI] [PubMed] [Google Scholar]
- Franssen F, Gerard C, Cozma‐Petruţ A, Vieira‐Pinto M, Jambrak AR, Rowan N, Paulsen P, Rozycki M, Tysnes K and Rodriguez‐Lazaro D, 2019. Inactivation of parasite transmission stages: efficacy of treatments on food of animal origin. Trends in Food Science & Technology, 83, 114–128. [Google Scholar]
- Franssen F, Deng HF, Swart A, Marinovic AB, Liu XL, Liu MY and van der Giessen J, 2021. Inactivation of Trichinella muscle larvae at different time‐temperature heating profiles simulating home‐cooking. Experimental Parasitology, 224, 108099. 10.1016/j.exppara.2021.108099 [DOI] [PubMed] [Google Scholar]
- Gabriel S, Dorny P, Saelens G and Dermauw V, 2023. Foodborne parasites and their complex life cycles challenging food safety in different food chains. Foods, 12. 10.3390/foods12010142 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gallardo C, Okoth E, Pelayo V, Anchuelo R, Martin E, Simon A, Llorente A, Nieto R, Soler A, Martin R, Arias M and Bishop RP, 2011. African swine fever viruses with two different genotypes, both of which occur in domestic pigs, are associated with ticks and adult warthogs, respectively, at a single geographical site. Journal of General Virology, 92, 432–444. [DOI] [PubMed] [Google Scholar]
- Gardinali NR, Barry AF, Otonel RAA, Alfieri AF and Alfieri AA, 2012. Hepatitis E virus in liver and bile samples from slaughtered pigs of Brazil. Memorias Do Instituto Oswaldo Cruz, 107, 935–939. 10.1590/s0074-02762012000700016 [DOI] [PubMed] [Google Scholar]
- Garre A, Zwietering MH and den Besten HM, 2023. The importance of what we cannot observe: experimental limitations as a source of bias for meta‐regression models in predictive microbiology. International Journal of Food Microbiology, 387, 110045. [DOI] [PubMed] [Google Scholar]
- Gemmell DK, Mack A, Wegmann S, Han D, Tuccelli R, Johnson M and Miller C, 2021. Efficacy of minute virus of mice (MVM) inactivation utilizing high temperature short time (HTST) pasteurization and suitability assessment of pasteurized, concentrated glucose feeds in Chinese hamster ovary (CHO) cell expression systems. Engineering in Life Sciences, 21, 502–513. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Geng YS, Zhao CY, Guo TY, Xu Y, Wang XP, Huang WJ, Liu H and Wang YC, 2019. Detection of Hepatitis E virus in raw pork and pig viscera as food in Hebei province of China. Foodborne Pathogens and Disease, 16, 325–330. 10.1089/fpd.2018.2572 [DOI] [PubMed] [Google Scholar]
- Ghosh S, Soto E, Illanes O, Navarro R, Aung MS, Malik YS, Kobayashi N and Fuentealba C, 2018. High detection rates of Torque teno sus virus in co‐infection with important viral pathogens in porcine kidneys on St. Kitts Island, Lesser Antilles. Transboundary and Emerging Diseases, 65, 1175–1181. 10.1111/tbed.12960 [DOI] [PubMed] [Google Scholar]
- Gröner A, Broumis C, Fang R, Nowak T, Popp B, Schäfer W and Roth NJ, 2018. Effective inactivation of a wide range of viruses by pasteurization. Transfusion, 58, 41–51. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gutiérrez‐Vergara C, Quintero J, Duarte J, Suescún J and López‐Herrera A, 2015. Detection of hepatitis E virus genome in pig livers in antioquia, Colombia. Genetic Molecular Research, 14, 2890–2899. [DOI] [PubMed] [Google Scholar]
- Haby MM, Leon LAS, Lucianez A, Nicholls RS, Reveiz L and Donadeu M, 2020. Systematic review of the effectiveness of selected drugs for preventive chemotherapy for Taenia solium taeniasis. Plos Neglected Tropical Diseases, 14. 10.1371/journal.pntd.0007873 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Halaihel N, Masia RM, Fernandez‐Jimenez M, Ribes JM, Montava R, de Blas I, Girones O, Alonso JL and Buesa J, 2010. Enteric calicivirus and rotavirus infections in domestic pigs. Epidemiology and Infection, 138, 542–548. 10.1017/s0950268809990872 [DOI] [PubMed] [Google Scholar]
- Hause B, Duff JW, Scheidt A and Anderson G, 2016a. Virus detection using metagenomic sequencing of swine nasal and rectal swabs. Journal of Swine Health and Production, 24, 304–308. [Google Scholar]
- Hause BM, Myers O, Duff J and Hesse RA, 2016b. Senecavirus A in pigs, United States, 2015. Emerging Infectious Diseases, 22, 1323–1325. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hillen S, von Berg S, Kohler K, Reinacher M, Willems H and Reiner G, 2014. Occurrence and severity of lung lesions in slaughter pigs vaccinated against Mycoplasma hyopneumoniae with different strategies. Preventive Veterinary Medicine, 113, 580–588. [DOI] [PubMed] [Google Scholar]
- Ho S, Jun Y, Park C, Lee C and Bae J, 1999. Prevalence of tissue antigen and serum antibody for porcine reproductive and respiratory syndrome in Cheju. Korean Journal of Veterinary Research, 39, 760–764. [Google Scholar]
- Hu XF, Chen Z, Li Y, Ding Z, Zeng QH, Wan T and Wu HS, 2022. Detection of porcine circovirus 1/2/3 and genetic analysis of porcine circovirus 2 in wild boar from Jiangxi province of China. Animals, 12, 11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huang JH, Li Y, Liu MM, Xia YH and Li ZR, 2013. A novel subgenotype of Torque teno Virus 1 (TTSuV1) in Slaughter Pigs in China. Food and Environmental Virology, 5, 226–230. 10.1007/s12560-013-9126-0 [DOI] [PubMed] [Google Scholar]
- Huangfu C, Ma Y, Jia J, Lv M, Zhu F, Ma X, Zhao X and Zhang J, 2017. Inactivation of viruses by pasteurization at 60° C for 10 h with and without 40% glucose as stabilizer during a new manufacturing process of α2‐Macroglobulin from Cohn Fraction IV. Biologicals, 46, 139–142. [DOI] [PubMed] [Google Scholar]
- Intharasongkroh D, Sa‐nguanmoo P, Tuanthap S, Thongmee T, Duang‐in A, Klinfueng S, Chansaenroj J, Vongpunsawad S, Theamboonlers A, Payungporn S, Chirathaworn C and Poovorawan Y, 2017. Hepatitis E virus in pork and variety meats sold in fresh markets. Food and Environmental Virology, 9, 45–53. 10.1007/s12560-016-9258-0 [DOI] [PubMed] [Google Scholar]
- Jansen F, Dorny P, Gabriël S, Dermauw V, Johansen MV and Trevisan C, 2021. The survival and dispersal of Taenia eggs in the environment: what are the implications for transmission? A systematic review. Parasites and Vectors, 14, 1–16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jia YF, Zhu QL, Xu T, Chen XM, Li HX, Ma MY, Zhang YB, He ZJ and Chen HY, 2022. Detection and genetic characteristics of porcine circovirus type 2 and 3 in Henan of China. Molecular and Cellular Probes, 61, 8. [DOI] [PubMed] [Google Scholar]
- Jiang JF, Zha YF, Liu J, Xing CN, Mi SJ, Yu JX, Sun YW, Tu CC, Gong WJ and Lu ZJ, 2021. Isolation and evolutionary analysis of Senecavirus A isolates from Guangdong province, China. Infection Genetics and Evolution, 91, 7. [DOI] [PubMed] [Google Scholar]
- John JK, Das T, Sethi M, Kattoor JJ, Tomar N and Saikumar G, 2022. Epidemiological study of porcine teschovirus infection in pigs at Bareilly, Uttar Pradesh, India. Biological Rhythm Research, 53, 50–57. [Google Scholar]
- Jones TH and Johns MW, 2012. Assessment of F‐RNA Coliphage as a potential indicator of enteric virus contamination of hog carcasses. Journal of Food Protection, 75, 1492–1500. [DOI] [PubMed] [Google Scholar]
- Jones TH and Muehlhauser V, 2017a. F‐coliphages, porcine adenovirus and porcine teschovirus as potential indicator viruses of fecal contamination for pork carcass processing. International Journal of Food Microbiology, 241, 237–243. [DOI] [PubMed] [Google Scholar]
- Jones TH and Muehlhauser V, 2017b. Frequency of hepatitis E virus, rotavirus and porcine enteric calicivirus at various stages of pork carcass processing in two pork processing plants. International Journal of Food Microbiology, 259, 29–34. [DOI] [PubMed] [Google Scholar]
- Jori F, Laval M, Maestrini O, Casabianca F, Charrier F and Pavio N, 2016. Assessment of domestic pigs, wild boars and feral hybrid pigs as reservoirs of hepatitis E virus in Corsica, France. Viruses‐Basel, 8, 11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kadoi K, Iwabuchi M, Satoh T, Katase T, Kawaji T and Morichi T, 1997. Adenovirus isolation from spleen lymphocytes of apparently healthy pigs. Microbiologica, 20, 215–220. [PubMed] [Google Scholar]
- Kekarainen T and Segalés J, 2012. Torque teno sus virus in pigs: an emerging pathogen? Transboundary and Emerging Diseases, 59, 103–108. [DOI] [PubMed] [Google Scholar]
- Khounvisith V, Tritz S, Khenkha L, Phoutana V, Keosengthong A, Pommasichan S, Nouanthong P, Hubschen JM, Snoeck CJ, Reinharz D, Muller CP, Black AP and Pauly M, 2018. High circulation of Hepatitis E virus in pigs and professionals exposed to pigs in Laos. Zoonoses and Public Health, 65, 1020–1026. 10.1111/zph.12520 [DOI] [PubMed] [Google Scholar]
- Kleymann A, Soto E, Illanes O, Malik YS, Fuentealba C and Ghosh S, 2020. High rates of detection and complete genomic analysis of porcine circovirus 2 (PCV2) in the Lesser Antilles Island of St. Kitts: identification of PCV2b‐PCV2d recombinants. Transboundary and Emerging Diseases, 67, 2282–2289. 10.1111/tbed.13583 [DOI] [PubMed] [Google Scholar]
- Kong V, Sum P, Mol P, Chan K, Ratha S, Laut S and Venn V, 2021. Risk assessment of African swine fever virus in pork in Phnom Penh, Cambodia. International Journal of Environmental and Rural Development, 11, 146–152. [Google Scholar]
- Kwon TY, Lee SS, Kim CY, Shin JY, Sunwoo SY and Lyoo YS, 2011. Genetic characterization of H7N2 influenza virus isolated from pigs. Veterinary Microbiology, 153, 393–397. [DOI] [PubMed] [Google Scholar]
- Laconi A, Cavicchio L, Tassoni L, Cunial G, Milani A, Ustulin M, Di Martino G, Forzan M, Campalto M, Monne I and Beato MS, 2020. Identification of two divergent swine Noroviruses detected at the slaughterhouse in North East Italy. Porcine Health Management, 6, 6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lainšček PR, Toplak I and Kirbis A, 2017. A comprehensive study of hepatitis E virus infection in pigs entering a slaughterhouse in Slovenia. Veterinary Microbiology, 212, 52–58. [DOI] [PubMed] [Google Scholar]
- Laisse CJ, Souza CK, Pereira PR, De Lorenzo C, Bianchi MV, Mapaco LP, Pavarini SP, Canal CW and Driemeier D, 2018. Detection and phylogenetic characterization of porcine circovirus 2 from pigs in Mozambique. Journal of Veterinary Diagnostic Investigation, 30, 342–347. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lau SKP, Woo PCY, Yip CCY, Li KSM, Fu CTY, Huang Y, Chan KH and Yuen KY, 2011. Co‐existence of multiple strains of two novel porcine bocaviruses in the same pig, a previously undescribed phenomenon in members of the family Parvoviridae, and evidence for inter‐ and intra‐host genetic diversity and recombination. Journal of General Virology, 92, 2047–2059. [DOI] [PubMed] [Google Scholar]
- Leblanc D, Ward P, Gagne MJ, Poitras E, Muller P, Trottier YL, Simard C and Houde A, 2007. Presence of hepatitis E virus in a naturally infected swine herd from nursery to slaughter. International Journal of Food Microbiology, 117, 160–166. [DOI] [PubMed] [Google Scholar]
- Leblanc D, Poitras E, Gagne MJ, Ward P and Houde A, 2010. Hepatitis E virus load in swine organs and tissues at slaughterhouse determined by real‐time RT‐PCR. International Journal of Food Microbiology, 139, 206–209. [DOI] [PubMed] [Google Scholar]
- Leblanc D, Houde A, Gagne MJ, Plante D, Bellon‐Gagnon P, Jones TH, Muehlhauser V, Wilhelm B, Avery B, Janecko N and Brassard J, 2014. Presence, viral load and characterization of Torque teno sus viruses in liver and pork chop samples at retail. International Journal of Food Microbiology, 178, 60–64. [DOI] [PubMed] [Google Scholar]
- Lelie P, Reesink H and Lucas C, 1987. Inactivation of 12 viruses by heating steps applied during manufacture of a hepatitis B vaccine. Journal of Medical Virology, 23, 297–301. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li JH and McClane BA, 2008. A novel small acid soluble protein variant is important for spore resistance of most Clostridium perfringens food poisoning isolates. Plos Pathogens, 4, 11. 10.1371/journal.ppat.1000056 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li WG, Shu XH, Pu YL, Bi JL, Yang GS and Yin GF, 2011. Seroprevalence and molecular detection of hepatitis E virus in Yunnan Province, China. Archives of Virology, 156, 1989–1995. 10.1007/s00705-011-1089-6 [DOI] [PubMed] [Google Scholar]
- Li Z, Wang W, Liu D and Guo Y, 2018. Effects of Lactobacillus acidophilus on the growth performance and intestinal health of broilers challenged with Clostridium perfringens . Journal of Animal Science and Biotechnology, 9, 1–10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu MM, Li Y, Sun D, Xia YH, Huang JH and Guo LL, 2014. Detection and genetic analysis of porcine bocavirus in different swine herds in North Central China. Scientific World Journal, 7, 1–7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu JX, Zha YF, Li HZ, Sun YW, Wang FG, Lu R and Ning ZY, 2019. Novel recombinant Seneca valley virus isolated from slaughtered pigs in Guangdong province. Virologica Sinica, 34, 722–724. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Luka PD, Erume J, Yakubu B, Owolodun OA, Shamaki D and Mwiine FN, 2016. Molecular detection of Torque Teno Sus virus and coinfection with African swine fever virus in blood samples of pigs from some slaughterhouses in Nigeria. Advances in Virology, 2016, 6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lund BM and Peck MW, 2001. Clostridium botulinum. In: Labbé RG and Garcia S (eds). Guide to Foodborne Pathogens. pp. 69–85. [Google Scholar]
- Lund B, Jensen VF, Have P and Ahring B, 1996. Inactivation of virus during anaerobic digestion of manure in laboratory scale biogas reactors. Antonie van Leeuwenhoek, 69, 25–31. [DOI] [PubMed] [Google Scholar]
- Luo ZY, Roi S, Dastor M, Gallice E, Laurin MA and L'Homme Y, 2011. Multiple novel and prevalent astroviruses in pigs. Veterinary Microbiology, 149, 316–323. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ma M, Li J and McClane BA, 2012. Genotypic and phenotypic characterization of Clostridium perfringens isolates from Darmbrand cases in post‐World War II Germany. Infect Immunology, 80, 4354–4363. 10.1128/IAI.00818-12 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Machnowska P, Ellerbroek L and Johne R, 2014. Detection and characterization of potentially zoonotic viruses in faeces of pigs at slaughter in Germany. Veterinary Microbiology, 168, 60–68. [DOI] [PubMed] [Google Scholar]
- Magar R and Larochelle R, 2004. Evaluation of the presence of porcine reproductive and respiratory syndrome virus in pig meat and experimental transmission following oral exposure. Canadian Journal of Veterinary Research‐Revue Canadienne De Recherche Veterinaire, 68, 259–266. [PMC free article] [PubMed] [Google Scholar]
- Maheshwari G, Jannat R, McCormick L and Hsu D, 2004. Thermal inactivation of adenovirus type 5. Journal of Virological Methods, 118, 141–146. [DOI] [PubMed] [Google Scholar]
- Martinez‐Guino L, Kekarainen T, Maldonado J, Aramouni M, Llorens A and Segales J, 2010. Torque tenosus virus (TTV) detection in aborted and slaughterhouse collected foetuses. Theriogenology, 74, 277–281. [DOI] [PubMed] [Google Scholar]
- Masana MO and Baranyi J, 2000. Adding new factors to predictive models: the effect on the risk of extrapolation. Food Microbiology, 17, 367–374. [Google Scholar]
- McDonnell GE, 2020. Antisepsis, Disinfection, and Sterilization: Types, Action, and Resistance. John Wiley & Sons. [Google Scholar]
- Mengeling WL, Lager KM, Zimmerman JK, Samarikermani N and Beran GW, 1991. A current assessment of the role of porcine parvovirus as a cause of fetal porcine death. Journal of Veterinary Diagnostic Investigation, 3, 33–35. [DOI] [PubMed] [Google Scholar]
- Meseko C, Globig A, Ijomanta J, Joannis T, Nwosuh C, Shamaki D, Harder T, Hoffman D, Pohlmann A, Beer M, Mettenleiter T and Starick E, 2018. Evidence of exposure of domestic pigs to highly pathogenic avian influenza H5N1 in Nigeria. Scientific Reports, 8, 9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Milojević L, Velebit B, Teodorovic V, Kirbis A, Petrovic T, Karabasil N and Dimitrijevic M, 2019. Screening and molecular characterization of Hepatitis E Virus in slaughter pigs in Serbia. Food and Environmental Virology, 11, 410–419. 10.1007/s12560-019-09393-1 [DOI] [PubMed] [Google Scholar]
- Monini M, Vignolo E, Ianiro G, Ostanello F, Ruggeri FM and Di Bartolo I, 2016. Detection of Torque Teno Sus virus in pork bile and liver sausages. Food and Environmental Virology, 8, 283–288. 10.1007/s12560-016-9249-1 [DOI] [PubMed] [Google Scholar]
- Motoya T, Umezawa M, Goto K, Doi I, Nagata N, Ikeda Y, Sakuta A, Sasaki N and Ishii K, 2019. High prevalence of hepatitis E virus infection among domestic pigs in Ibaraki Prefecture, Japan. BMC Veterinary Research, 15, 8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mughini‐Gras L, Angeloni G, Salata C, Vonesch N, D'Amico W, Campagna G, Natale A, Zuliani F, Ceglie L, Monne I, Vascellari M, Capello K, Di Martino G, Inglese N, Palu G, Tomao P and Bonfanti L, 2017. Hepatitis E virus infection in North Italy: high seroprevalence in swine herds and increased risk for swine workers. Epidemiology and Infection, 145, 3375–3384. 10.1017/s0950268817002485 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Muller A, Collineau L, Stephan R, Muller A and Stark KDC, 2017. Assessment of the risk of foodborne transmission and burden of hepatitis E in Switzerland. International Journal of Food Microbiology, 242, 107–115. [DOI] [PubMed] [Google Scholar]
- Ng H, Bayne HG and Garibaldi JA, 1969. Heat resistance of Salmonella: the uniqueness of Salmonella senftenberg 775W. Applied Microbiology, 17, 78–82. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nims R and Plavsic M, 2013. Intra‐family and inter‐family comparisons for viral susceptibility to heat inactivation. Journal of Microbial and Biochemical Technology, 5, 136–141. [Google Scholar]
- Nims RW and Zhou SS, 2016. Intra‐family differences in efficacy of inactivation of small, non‐enveloped viruses. Biologicals, 44, 456–462. [DOI] [PubMed] [Google Scholar]
- Nishizawa T, Okamoto H, Konishi K, Yoshizawa H, Miyakawa Y and Mayumi M, 1997. A novel DNA virus (TTV) associated with elevated transaminase levels in posttransfusion hepatitis of unknown etiology. Biochemical and Biophysical Research Communications, 241, 92–97. [DOI] [PubMed] [Google Scholar]
- Nokireki T, Laine T, London L, Ikonen N and Huovilainen A, 2013. The first detection of influenza in the Finnish pig population: a retrospective study. Acta Veterinaria Scandinavica, 55, 7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Novak JS and Yuan JTC, 2003. Viability of Clostridium perfringens, Escherichia coli, and listeria monocytogenes surviving mild heat or aqueous ozone treatment on beef followed by heat, alkali, or salt stress. Journal of Food Protection, 66, 382–389. 10.4315/0362-028x-66.3.382 [DOI] [PubMed] [Google Scholar]
- O'Dea MA, Hughes AP, Davies LJ, Muhling J, Buddle R and Wilcox G, 2008. Thermal stability of porcine circovirus type 2 in cell culture. Journal of Virological Methods, 147, 61–66. [DOI] [PubMed] [Google Scholar]
- Ojok L, Okuni JB, Hohloch C, Hecht W and Reinacher M, 2013. Detection and characterisation of Porcine Circovirus 2 from Ugandan pigs. Indian Journal of Veterinary Pathology, 37, 77–80. [Google Scholar]
- Oladipo EK, Adeniji JA and Fagbami AH, 2013. Isolation of influenza A and B viruses from pigs at Bodija abattoir, Ibadan, Nigeria. Global Veterinaria, 11, 252–257. [Google Scholar]
- Olaniyi MO, Adebiyi AA, Ajayi OL, Alaka OO and Akpavie SO, 2020. Localization and immunohistochemical detection of swine influenza A virus subtype H1N1 antigen in formalin‐fixed, paraffin‐embedded lung tissues from naturally infected pigs. Beni‐Suef University Journal of Basic and Applied Sciences, 9, 8. [Google Scholar]
- Orsburn B, Melville SB and Popham DL, 2008. Factors contributing to heat resistance of Clostridium perfringens endospores. Applied Environmental Microbiology, 74, 3328–3335. 10.1128/AEM.02629-07 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Osoro EM, Lidechi S, Nyaundi J, Marwanga D, Mwatondo A, Muturi M, Ng'ang'a Z and Njenga K, 2019. Detection of pandemic influenza A/H1N1/pdm09 virus among pigs but not in humans in slaughterhouses in Kenya, 2013‐2014. BMC Research Notes, 12, 6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- O'Sullivan T, Friendship R, Blackwell T, Pearl D, McEwen B, Carman S, Slavic D and Dewey C, 2011. Microbiological identification and analysis of swine tonsils collected from carcasses at slaughter. Canadian Journal of Veterinary Research‐Revue Canadienne De Recherche Veterinaire, 75, 106–111. [PMC free article] [PubMed] [Google Scholar]
- Owolodun OA, Yakubu B, Antiabong JF, Adefalujo OK, Ogedengbe ME, Obisakin ET and Shamaki D, 2007. Molecular assessment of African swine fever in North‐Central Nigeria. Bulletin of Animal Health and Production in Africa, 55, 96–103. [Google Scholar]
- Owolodun OA, Obishakin ET, Ekong PS and Yakubu B, 2010. Investigation of African swine fever in slaughtered pigs, Plateau state, Nigeria, 2004–2006. Tropical Animal Health and Production, 42, 1605–1610. [DOI] [PubMed] [Google Scholar]
- Paladino ES, Gabardo MDP, Lunardi PN, Mores N and Guedes RMC, 2017. Anatomopathological pneumonic aspects associated with highly pathogenic Pasteurella multocida in finishing pigs. Pesquisa Veterinaria Brasileira, 37, 1091–1100. 10.1590/s0100-736x2017001000009 [DOI] [Google Scholar]
- Paluszak Z, Lipowski A and Ligocka A, 2010. Survival of BPV and Aujeszky's disease viruses in meat wastes subjected to different sanitization processes. Polish Journal of Veterinary Sciences, 13, 749–753. [DOI] [PubMed] [Google Scholar]
- Papatsiros VG, Athanasiou LV, Psalla D, Petridou E, Maragkakis GG, Papatsas I, Arsenakis I and Maes D, 2015. Erythema multiforme associated with respiratory disease in a commercial breeding pig herd. Viral Immunology, 28, 464–471. [DOI] [PubMed] [Google Scholar]
- Paredes‐Sabja D, Sarker N, Setlow B, Setlow P and Sarker MR, 2008. Roles of dacB and spm proteins in Clostridium perfringens spore resistance to moist heat, chemicals, and UV radiation. Applied and Environmental Microbiology, 74, 3730–3738. 10.1128/aem.00169-08 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Park JY, Welch MW, Harmon KM, Zhang JQ, Pineyro PE, Li GW, Hause BM and Gauger PC, 2019. Detection, isolation, and in vitro characterization of porcine parainfluenza virus type 1 isolated from respiratory diagnostic specimens in swine. Veterinary Microbiology, 228, 219–225. [DOI] [PubMed] [Google Scholar]
- Peleg M, 2021. The thermal death time concept and its implications revisited. Food Engineering Reviews, 13, 291–303. [Google Scholar]
- Pellerin M, Trabucco B, Capai L, Laval M, Maestrini O, Jori F, Falchi A, Doceul V, Charrier F, Casabianca F and Pavio N, 2022. Low prevalence of hepatitis E virus in the liver of Corsican pigs slaughtered after 12 months despite high antibody seroprevalence. Transboundary and Emerging Diseases, 69, E2706–E2718. 10.1111/tbed.14621 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Perera HKK, Wickramasinghe G, Cheung CL, Nishiura H, Smith DK, Poon LLM, Perera AKC, Ma SK, Sunil‐Chandra NP, Guan Y and Peiris JSM, 2013. Swine influenza in Sri Lanka. Emerging Infectious Diseases, 19, 481–484. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Peter M and Kühnel F, 2020. Oncolytic adenovirus in cancer immunotherapy. Cancers, 12, 3354. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pogranichniy R, Lee K and Machaty Z, 2008. Detection of porcine parvovirus in the follicular fluid of abattoir pigs. Journal of Swine Health and Production, 16, 244–246. [Google Scholar]
- Pourcher AM, Devriese L, Hernandez J and Delattre J, 1991. Enumeration by a miniaturized method of Escherichia coli, Streptococcus bovis and enterococci as indicators of the origin of faecal pollution of waters. Journal of Applied Bacteriology, 70, 525–530. [DOI] [PubMed] [Google Scholar]
- Qiao CL, Liu LP, Yang HL, Chen Y, Xu HY and Chen HL, 2014. Novel triple reassortant H1N2 influenza viruses bearing six internal genes of the pandemic 2009/H1N1 influenza virus were detected in pigs in China. Journal of Clinical Virology, 61, 529–534. [DOI] [PubMed] [Google Scholar]
- Quintana J, Segales J, Rosell C, Calsamiglia M, Rodriguez‐Arrioja GM, Chianini F, Folch JM, Maldonado J, Canal M, Plana‐Duran J and Domingo M, 2001. Clinical and pathological observations on pigs with postweaning multisystemic wasting syndrome. Veterinary Record, 149, 357–361. [DOI] [PubMed] [Google Scholar]
- Raju D and Sarker MR, 2005. Comparison of the levels of heat resistance of wild‐type, cpe knockout, and cpe plasmid‐cured Clostridium perfringens type A strains. Applied and Environmental Microbiology, 71, 7618–7620. 10.1128/aem.71.11.7618-7620.2005 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Raju D and Sarker MR, 2007. Production of small, acid‐soluble spore proteins in Clostridium perfringens nonfoodborne gastrointestinal disease isolates. Canadian Journal of Microbiology, 53, 514–518. 10.1139/w07-016 [DOI] [PubMed] [Google Scholar]
- R Core Team (2021). R: A language and environment for statistical computing. R Foundation for Statistical Computing, Vienna, Austria. URL https://www.R–project.org/.
- Rose N, Lunazzi A, Dorenlor V, Merbah T, Eono F, Eloit M, Madec F and Pavio N, 2011. High prevalence of Hepatitis E virus in French domestic pigs. Comparative Immunology Microbiology and Infectious Diseases, 34, 419–427. [DOI] [PubMed] [Google Scholar]
- Rose N, Herve S, Eveno E, Barbier N, Eono F, Dorenlor V, Andraud M, Camsusou C, Madec F and Simon G, 2013. Dynamics of influenza A virus infections in permanently infected pig farms: evidence of recurrent infections, circulation of several swine influenza viruses and reassortment events. Veterinary Research, 44, 14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rosnes J, Fernandez P, Periago P and Shara T, 2012. Microorganisms of relevance in thermally processed foods. Progress on Quantitative Approaches of Thermal Food Processing. Nova Science Publishers, New York. pp. 1–37. [Google Scholar]
- Rout M and Saikumar G, 2016. Development and application of real‐time TagMan RT‐PCR assay for improved detection of classical swine fever virus in slaughtered pigs. Indian Journal of Animal Research, 50, 979–982. [Google Scholar]
- Rout M, Saikumar G and Nagarajan K, 2015. Diagnostic potential of polymerase chain reaction in detection of classical swine fever virus infection in slaughtered pigs. Indian Journal of Animal Research, 49, 512–514. [Google Scholar]
- Saekhow P, Kishizuka S, Sano N, Mitsui H, Akasaki H, Mawatari T and Ikeda H, 2015. Coincidental detection of genomes of porcine parvoviruses and porcine circovirus type 2 infecting pigs in Japan. Journal of Veterinary Medical Science, 77, 1581–1586. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sahlström L, 2003. A review of survival of pathogenic bacteria in organic waste used in biogas plants. Bioresource Technology, 87, 161–166. [DOI] [PubMed] [Google Scholar]
- Sahlström L, Bagge E, Emmoth E, Holmqvist A, Danielsson‐Tham M‐L and Albihn A, 2008. A laboratory study of survival of selected microorganisms after heat treatment of biowaste used in biogas plants. Bioresource Technology, 99, 7859–7865. [DOI] [PubMed] [Google Scholar]
- Sarker MR, Shivers RP, Sparks SG, Juneja VK and McClane BA, 2000. Comparative experiments to examine the effects of heating on vegetative cells and spores of Clostridium perfringens isolates carrying plasmid genes versus chromosomal enterotoxin genes. Applied and Environmental Microbiology, 66, 3234–3240. 10.1128/aem.66.8.3234-3240.2000 [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- Sarma DK and Meshram DJ, 2008. Comparison of sandwich and dot ELISA for detection of CSF virus antigen in pigs. Indian Veterinary Journal, 85, 915–918. [Google Scholar]
- Sarma DK, Sarma S, Tiwari AK, Rajkhowa S, Meshram DJ, Singh NK and Baruah KK, 2007. Occurrence of classical swine fever virus in tissues of slaughtered pigs in different districts of Assam. Indian Journal of Virology, 18, 61–64. [Google Scholar]
- Sarma DK, Krishna L and Mishri J, 2008. Classical swine fever in pigs and its status in India: A review. Indian Journal of Animal Sciences, 78, 1311–1317. [Google Scholar]
- Saucier L and Plamondon É, 2011. Heat inactivation of Mycobacterium avium subsp. paratuberculosis in aseptically prepared ground beef. International Journal of Food Engineering, 7. [Google Scholar]
- Sauerbrei A and Wutzler P, 2009. Testing thermal resistance of viruses. Archives of Virology, 154, 115–119. [DOI] [PubMed] [Google Scholar]
- Sharmila M and Sherikar AA, 2005. Detection and isolation of foot‐and‐mouth disease virus from fresh and frozen meats. Blue Cross Book: 41–48. [Google Scholar]
- Shen HG, Schalk S, Halbur PG, Campbell JM, Russell LE and Opriessnig T, 2011. Commercially produced spray‐dried porcine plasma contains increased concentrations of porcine circovirus type 2 DNA but does not transmit porcine circovirus type 2 when fed to naive pigs. Journal of Animal Science, 89, 1930–1938. 10.2527/jas.2010-3502 [DOI] [PubMed] [Google Scholar]
- Shirasaki N, Matsushita T, Matsui Y and Koriki S, 2020. Suitability of pepper mild mottle virus as a human enteric virus surrogate for assessing the efficacy of thermal or free‐chlorine disinfection processes by using infectivity assays and enhanced viability PCR. Water research, 186, 116409. [DOI] [PubMed] [Google Scholar]
- Soni A, Bremer P and Brightwell G, 2022. A comprehensive review of variability in the thermal resistance (D‐values) of food‐borne pathogens‐A challenge for thermal validation trials. Foods, 11, 18. 10.3390/foods11244117 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sooryanarain H, Heffron CL, Hill DE, Fredericks J, Rosenthal BM, Werre SR, Opriessnig T and Meng XJ, 2020. Hepatitis E Virus in Pigs from Slaughterhouses, United States, 2017–2019. Emerging Infectious Diseases, 26, 354–357. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sörqvist S, 2003. Heat resistance in liquids of Enterococcus spp., Listeria spp., Escherichia coli, Yersinia enterocolitica, Salmonella spp. and Campylobacter spp. Acta Veterinaria Scandinavica, 44, 1–19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Spillmann S, Traub F, Schwyzer M and Wyler R, 1987. Inactivation of animal viruses during sewage sludge treatment. Applied and Environmental Microbiology, 53, 2077–2081. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Streck AF, Homeier T, Foerster T, Fischer S and Truyen U, 2013. Analysis of porcine parvoviruses in tonsils and hearts from healthy pigs reveals high prevalence and genetic diversity in Germany. Archives of Virology, 158, 1173–1180. 10.1007/s00705-013-1603-0 [DOI] [PubMed] [Google Scholar]
- Swati K, Rahul S and Saikumar G, 2020. Epidemiological study of porcine sapelovirus infection in pigs at Bareilly area of Uttar Pradesh, India. Biological Rhythm Research, 51, 1155–1165. [Google Scholar]
- Tajima T and Kawamura H, 1998. Serological relationship among porcine cytomegalovirus Japanese isolates and a UK isolate. Journal of Veterinary Medical Science, 60, 107–109. [DOI] [PubMed] [Google Scholar]
- Takahashi M, Tanaka T, Takahashi H, Hoshino Y, Nagashima S, Jirintai F, Mizuo H, Mizuo Y, Yazaki T, Takagi T and Azuma M, 2010. Hepatitis E Virus (HEV) strains in serum samples can replicate efficiently in cultured cells despite the coexistence of HEV antibodies: characterization of HEV virions in blood circulation. Journal of Clinical Microbiology, 48, 1112–1125. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Takemae N, Harada M, Nguyen PT, Nguyen T, Nguyen TN, Nguyen TD, Pham VP, Le VT, Do HT, Vo HV, Le QVT, Tran TM, Nguyen TD, Thai PD, Nguyen DH, Le AQT, Nguyen DT, Uchida Y and Saito T, 2017. Influenza A viruses of Swine (IAV‐S) in Vietnam from 2010 to 2015: multiple introductions of A(H1N1)pdm09 Viruses into the pig population and diversifying genetic constellations of enzootic IAV‐S. Journal of Virology, 91, 18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Temmam S, Besnard L, Andriamandimby SF, Foray C, Rasamoelina‐Andriamanivo H, Heraud JM, Cardinale E, Dellagi K, Pavio N, Pascalis H and Porphyre V, 2013. High prevalence of Hepatitis E in humans and pigs and evidence of genotype‐3 virus in Swine, Madagascar. American Journal of Tropical Medicine and Hygiene, 88, 329–338. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thomas LF, Bishop RP, Onzere C, McIntosh MT, Lemire KA, e Glanville WA, EAJ C and Fevre EM, 2016. Evidence for the presence of African swine fever virus in an endemic region of Western Kenya in the absence of any reported outbreak. BMC Veterinary Research, 12, 10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thuy NTD, Trung NT, Dung TQ, Khoa DVA, Thuy DTN and Opriessnig T, 2021. First investigation of the prevalence of parvoviruses in slaughterhouse pigs and genomic characterization of ungulate copiparvovirus 2 in Vietnam. Archives of Virology, 166, 779–788. 10.1007/s00705-020-04928-5 [DOI] [PubMed] [Google Scholar]
- Traore KA, Ouoba JB, Huot N, Rogee S, Dumarest M, Traore AS, Pavio N, Barro N and Rogues P, 2015. Hepatitis E virus exposure is increased in pork butchers from Burkina Faso. American Journal of Tropical Medicine and Hygiene, 93, 1356–1359. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tuboly T and Nagy É, 2001. Construction and characterization of recombinant porcine adenovirus serotype 5 expressing the transmissible gastroenteritis virus spike gene. Journal of General Virology, 82, 183–190. [DOI] [PubMed] [Google Scholar]
- Tuladhar E, Bouwknegt M, Zwietering M, Koopmans M and Duizer E, 2012. Thermal stability of structurally different viruses with proven or potential relevance to food safety. Journal of Applied Microbiology, 112, 1050–1057. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ugwuanyi J, Harvey L and McNeil B, 1999. Effect of process temperature, pH and suspended solids content upon pasteurization of a model agricultural waste during thermophilic aerobic digestion. Journal of Applied Microbiology, 87, 387–395. [DOI] [PubMed] [Google Scholar]
- Vieira YR, os Santos DRL, Portilho MM, CEP V, Arissawa M, Villar LM, Pinto MA and e Paula VS, 2014. Hepadnavirus detected in bile and liver samples from domestic pigs of commercial abattoirs. Bmc Microbiology, 14, 6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vonlanthen‐Specker I, Stephan R, Sidler X, Moor D, Fraefel C and Bachofen C, 2021. Genetic diversity of Hepatitis E virus type 3 in Switzerland‐from stable to table. Animals, 11, 18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Walker L, Piela TH, Yates VJ and Gulka CM, 1982. Heat inactivation of a duck adenovirus serologically indistinguishable from adenovirus 127. Avian Diseases, 26, 602–605. [PubMed] [Google Scholar]
- Wang J, Zhang W, Ju H, Yang D, Liu J, Zhou J, Yuan S and Liu P, 2015. Investigation on the prevalence of porcine reproductive and respiratory syndrome in Shanghai area from 2004 to 2012. Zhongguo Yufang Shouyi Xuebao/Chinese Journal of Preventive Veterinary Medicine, 37, 190–193. [Google Scholar]
- Wang KR, Liu LB, Wang JF, Sun XX, Han QA, Zhou C, Xu XD and Wang JC, 2023. Quantification of hepatitis E virus in raw pork livers using droplet digital RT‐PCR. Food Microbiology, 109, 7. [DOI] [PubMed] [Google Scholar]
- Watcharasukarn M, Kaparaju P, Steyer J‐P, Krogfelt KA and Angelidaki I, 2009. Screening Escherichia coli, Enterococcus faecalis, and Clostridium perfringens as indicator organisms in evaluating pathogen‐reducing capacity in biogas plants. Microbial Ecology, 58, 221–230. [DOI] [PubMed] [Google Scholar]
- Welch J, Bienek C, Gomperts E and Simmonds P, 2006. Resistance of porcine circovirus and chicken anemia virus to virus inactivation procedures used for blood products. Transfusion, 46, 1951–1958. [DOI] [PubMed] [Google Scholar]
- Wen S, Sun W, Li Z, Zhuang X, Zhao G, Xie C, Zheng M, Jing J, Xiao P, Wang M, Han J, Ren J, Liu H, Lu H and Jin N, 2018. The detection of porcine circovirus 3 in Guangxi, China. Transboundary and Emerging Diseases, 65, 27–31. 10.1111/tbed.12754 [DOI] [PubMed] [Google Scholar]
- Wheeler AL, Hartel PG, Godfrey DG, Hill JL and Segars WI, 2002. Potential of Enterococcus faecalis as a human fecal indicator for microbial source tracking. Journal of Environmental Quality, 31, 1286–1293. [DOI] [PubMed] [Google Scholar]
- Winfield MD and Groisman EA, 2003. Role of nonhost environments in the lifestyles of Salmonella and Escherichia coli . Applied and Environmental Microbiology, 69, 3687–3694. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yue WD, Li YL, Zhang XR, He JP and Ma HL, 2022. Prevalence of porcine circoviruses in slaughterhouses in central Shanxi province, China. Frontiers in Veterinary Science, 9, 7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yunoki M, Tsujikawa M, Urayama T, Sasaki Y, Morita M, Tanaka H, Hattori S, Takechi K and Ikuta K, 2003. Heat sensitivity of human parvovirus B19. Vox Sanguinis, 84, 164–169. 10.1046/j.1423-0410.2003.00280.x [DOI] [PubMed] [Google Scholar]
- Zhang LH, Huang SC, Li K, Rehman MU, Jiang X, Tong XL, Zhang H, Iqbal MK, Mehmood K, Liu SZ, Shen YQ and Li JK, 2018. Molecular detection of indigenous Hepatitis E Virus (HEV) from Tibetan Pigs in Tibet, China. Food and Environmental Virology, 10, 373–377. 10.1007/s12560-018-9352-6 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Protocol for the assessment of the efficacy of methods 2 to 5 and method 7 to inactivate relevant pathogens when producing processed animal protein of porcine origin intended to feed poultry and aquaculture animals (EFSA‐Q‐2022‐00455)


