Figure 2.
Meteor depletion induces chromatin changes in mESCs
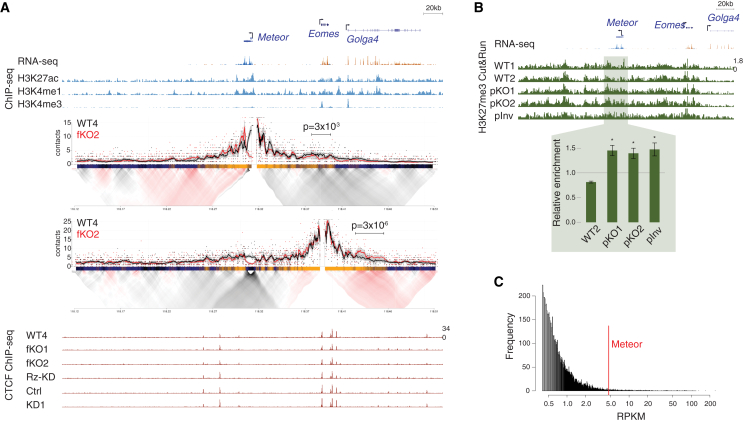
(A) (Top) Genome browser image of the region surrounding the Meteor locus. Shown are representative transcript models; RNA-seq tracks where orange denotes transcription on the plus strand and blue denotes transcription on the minus strand; and ENCODE mESC ChIP-seq tracks. (Middle) 4C analysis in the indicated mESC lines using the Meteor or Eomes promoters as viewpoints. Domainograms showing mean contact per fragment end for a series of window sizes are placed below smoothed trend lines and raw counts of the contact profiles. (Bottom) ChIP-seq tracks of CTCF in the indicated mESC lines. All tracks are normalized to the same scale.
(B) (Top) Genome browser image of the region surrounding the Meteor locus. RNA-seq track is the same as shown in (A); Cut&Run analysis of H3K27me3 levels in the indicated mESC lines grown in serum-free 2i/LIF conditions. (Bottom) Bar plot shows quantification of signal in the highlighted region, normalized to WT1 and to a H3K27me3-rich region near the Ppib gene (see STAR Methods). Bars represent standard errors; n = 3. ∗p < 0.05, one-sided t test.
(C) Distribution of reads per kilobase per million (RPKM) of all transcripts identified in an EZH2 RNA immunoprecipitation (RIP) dataset taken from Zhao et al.43 RPKM of Meteor indicated by a red line.
See also Figure S2.