Figure 5.
A distinct functional feature of the Meteor locus represses Eomes throughout neuronal differentiation
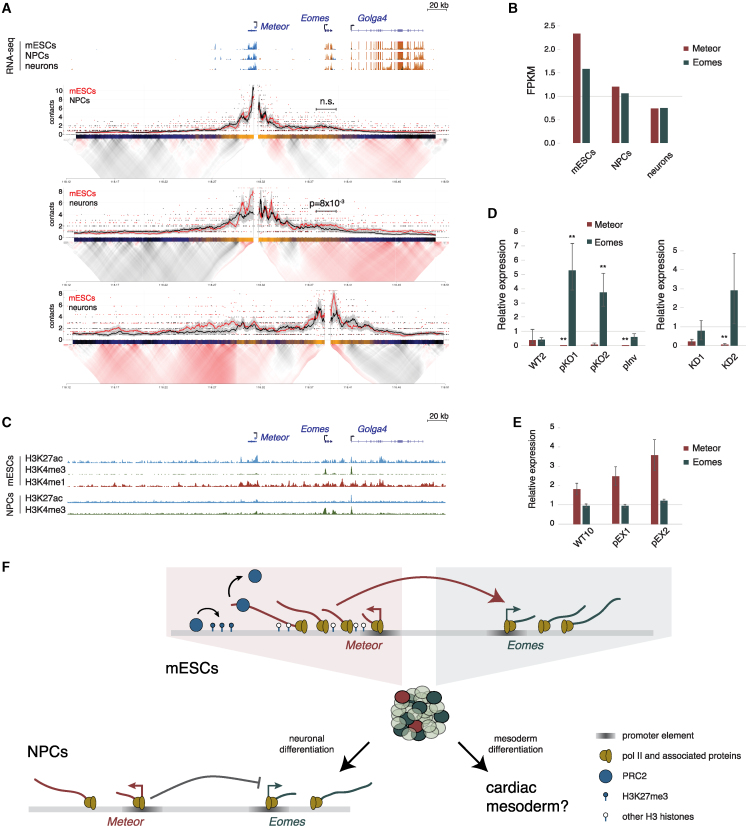
(A) (Top) Genome browser image of the region surrounding the Meteor locus. Shown are representative transcript models, and RNA-seq tracks taken from Hezroni et al.65 where orange denotes transcription on the plus strand and blue denotes transcription on the minus strand. All tracks are normalized to the same scale. (Bottom) 4C analysis in the indicated cells using the Meteor or Eomes promoters as viewpoints. Domainograms showing mean contact per fragment end for a series of window sizes are placed below smoothed trend lines and raw counts of the contact profiles.
(B) RSEM quantifications of Meteor and Eomes expression levels in the indicated cell types; RNA-seq data are the same as shown in (A).
(C) ENCODE ChIP-seq tracks in mESCs (top), and H3K27ac ChIP-seq and H3K4me3 Cut&Run tracks in NPCs (bottom). Genomic coordinates are aligned to (A).
(D) DESeq2 quantifications of Meteor and Eomes in NPCs derived from the indicated cell lines. Amounts normalized to WT1/Ctrl. Bars represent standard errors; n = 3. ∗∗padj < 0.005.
(E) qRT-PCR quantifications of Meteor and Eomes in NPCs derived from the indicated cell lines. Levels were normalized to WT9 and Ppib for internal control. Bars represent standard errors; n = 3.
(F) Model of Meteor function. In mESCs, the Meteor locus activates Eomes expression. Perturbing elongation through the locus is associated with increased H3K27me3 deposition and decreased Eomes expression, likely through decreasing the Eomes-expressing mESC subpopulation. As a function of the growth conditions of the cells, this might result in reduced efficiency of cardiac mesoderm formation. Following neuronal differentiation, the Meteor locus now represses Eomes levels, with the DNA element or transcription initiation serving as the functional feature.
See also Figure S5.