Fig. 2.
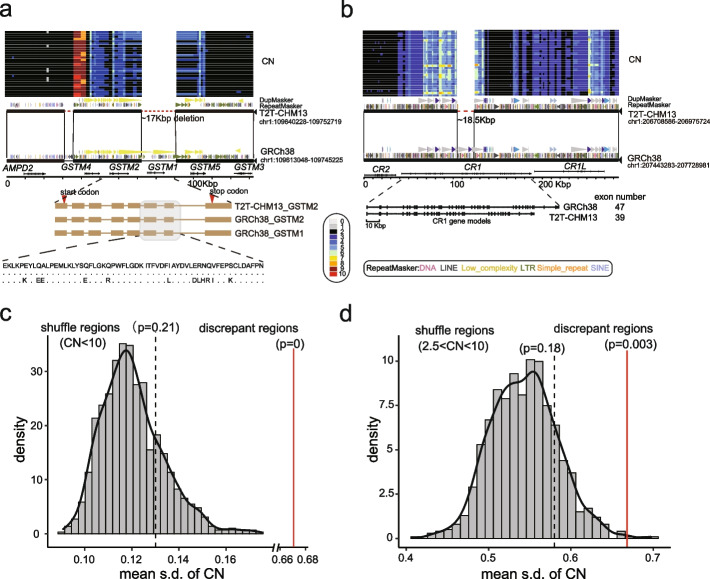
Gene structure differences in the discrepant regions. a The depletion of GSTM1 in the T2T-CHM13 genome assembly by a ~ 17 kbp deletion. The CN heatmap inferred from SGPD is shown in the top panel. The miropeat synteny relationship shows structural variation with repeat, duplication, and gene annotation. The exon schematic with amino acid alignment shows the gene-model difference in the two assemblies. b The depletion of eight exons of CR1 in T2T-CHM13 by ~ 18.5 kbp deletion. c The distribution of the mean of s.d. of CN shows the mean s.d. of 131 discrepant regions (mean = 0.735, red line) is significantly higher than the simulated null distribution of s.d. of CN (CN < 10, empirical p = 0). The black line represents the observed mean of s.d. of CN of the regions where the CN is less than 10. d The distribution of the mean s.d. of CN shows the mean s.d. of 131 discrepant regions (mean = 0.735, red line) is significantly higher than the simulated null distribution of s.d. of CN (2.5 < CN < 10, empirical p = 0). The black line represents the observed mean s.d. of CN of the regions where the CN is less than 10 and greater than 2.5