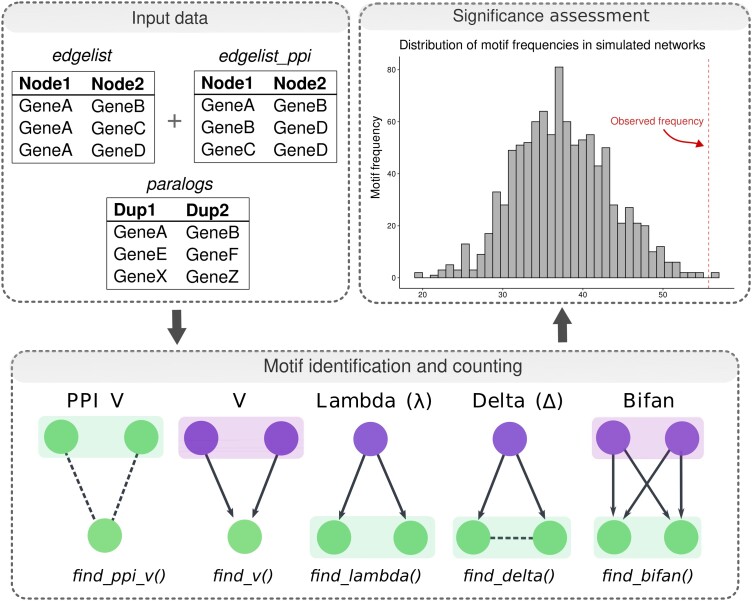
Fig. 1.
Workflow of motif identification and significance assessment with magrene. Input data types for motif detection, which include a data frame of GRN edges, a data frame of PPI network edges, and a data frame of paralogous gene pairs. Motif types and R functions in magrene to find specific motifs are indicated in the bottom panel. Shaded boxes indicate paralogous gene pairs. Regulators and targets are indicated in purple and green, respectively. Arrows indicate directed regulatory interactions, whereas dashed lines indicate protein–protein interactions. Observed motif frequencies for each motif type are then compared with a null distribution obtained from simulated networks, as indicated in the histogram. See Materials and Methods for details.