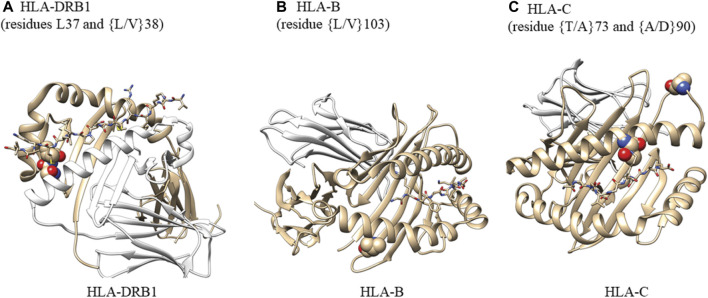
FIGURE 5.
Analysis of amino acid residues showing their potential ability to bind antigens. (A) Atoms in residues 37 and 38 are shown as spheres. HLA-DRA (cyan), HLA-DRB1 (gray), M141 TCR𝜶 (wheat), and M141 TCRβ (yellow) are shown as ribbons. Residues 37 and 38 are located on the peptide-binding surface. One of the two residues is also in contact with a helix from HLA-DRA that comprises the complex. We created this figure using PDB ID (4x5w); (B) Atoms in residues 103 are shown as spheres. HLA-B (gray) and β-2-macroglobulin (wheat) are shown as ribbons. The small peptide-binding pocket (lines) is composed of three helices, one of which directly interacts with residue 103 within HLA-B. This residue is present at the end of a b-sheet while in contact with a few hydrophobic residues from the helix above and may be important for maintaining the structural scaffold and creating the binding surface We created this figure using PDB ID (1K5N); (C) Atoms in residues 73 and 90 are shown as spheres. HLA-C (gray) and β-2-macroglobulin (wheat) are shown as ribbons. The small binding peptide is in direct contact with residue 73, and residue 90 is located in a loop region just after the helix present on the peptide-binding surface. We created this figure using PDB ID (6JT0).