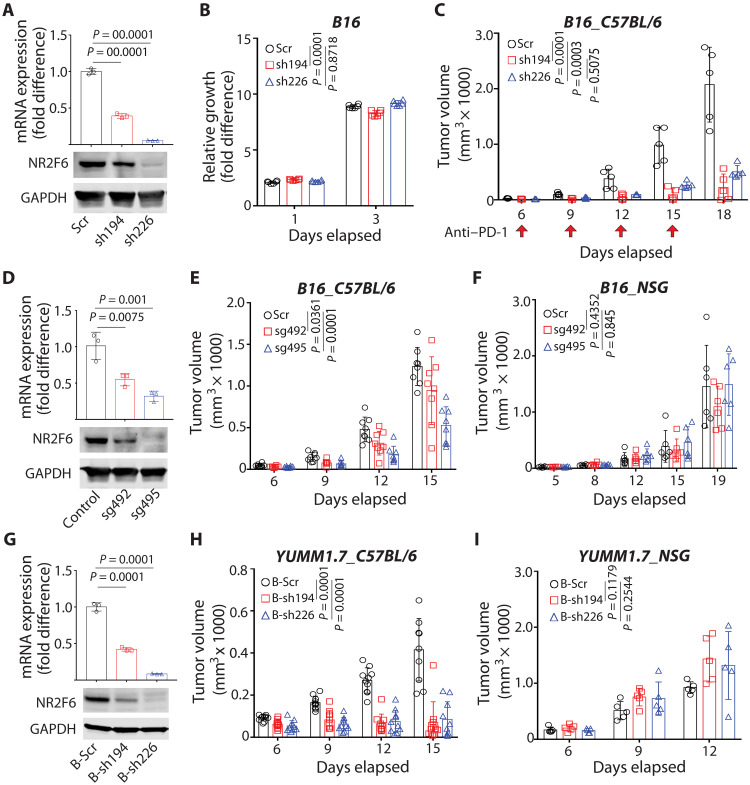
Fig. 2. NR2F6 control of murine melanoma growth requires an intact immune system.
(A) B16F10 cells were transduced with scramble (Scr) control shRNA or one of two shRNAs (sh194 and sh226) targeting murine Nr2f6. NR2F6 mRNA and protein expression was assessed by qPCR and immunoblotting. n = 3 for each group. GAPDH, glyceraldehyde-3-phosphate dehydrogenase. (B) Growth of cultured cells described in (A), as assessed in vitro using CellTiter-Glo. Relative fold differences in luminescence on days 1 and 3 were calculated relative to luminescence on day 0. n = 6 for each group. (C) C57BL/6 mice were then inoculated with cells and treated with anti–PD-1 antibody (RMP1-14) on days 6, 9, 12, and 15 (arrows). Tumor volumes were monitored at indicated time points. n = 5 mice for each group. (D) NR2F6 KO B16F10 cells were established by CRISPR using specific single guide RNAs (sg492 and sg495). NR2F6 mRNA and protein expression was assessed by qPCR and immunoblotting, respectively. n = 3 for each group. (E and F) Cells established in (D) were used to inoculate C57BL6 (E) or NSG (F) mice, and tumor volumes were monitored at indicated time points. n = 8 mice (E) and n = 6 mice (F) for each group. (G) YUMM1.7 cells were transduced with scrambled (B-Scr) control shRNA or one of two shRNAs (B-sh194 and B-sh226) targeting Nr2f6 as in (A). mRNA and protein expression was assessed by qPCR and immunoblotting, respectively. n = 3 for each group. (H and I) Cells established in (G) were then used to inoculate C57BL/6 (H) or NSG (I) mice, and tumor volumes were monitored at indicated time points. n = 9 mice (H) and n = 5 mice (I) for each group. Data are presented as means ± SD. Statistical significance was assessed by one-way analysis of variance (ANOVA) with Dunnett’s test (A, D, and G) and two-way ANOVA with Dunnett’s test (B, C, E, F, H, and I).