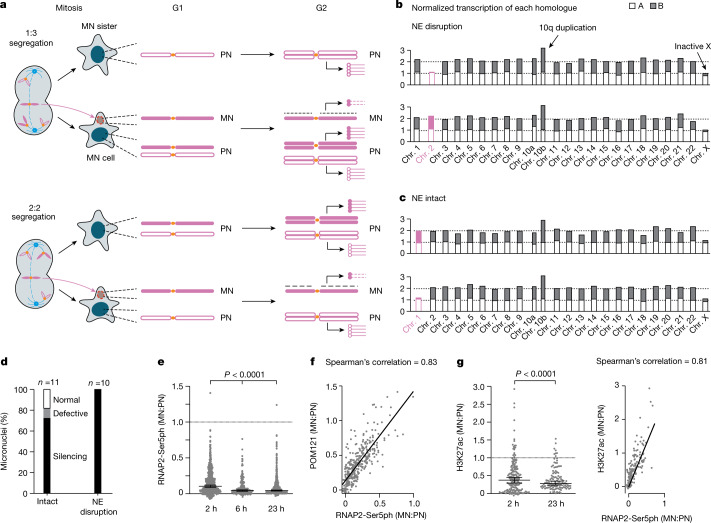
Fig. 1. Transcription defects in newly generated micronuclei.
a, Two patterns of chromosome segregation that generate a micronucleated cell (MN cell) and its sister (MN sister). Filled magenta shapes indicate the mis-segregated chromatid in the micronucleus (MN) and its sister chromatids in the primary nucleus (PN); open magenta shapes indicate sister chromatids of the other homologue. Top, a 1:3 mis-segregation generates monosomy in a MN sister cell and trisomy in a MN cell. Bottom, a 2:2 segregation generates disomy in both cells. In G2 (when cells were isolated), chromosomes in the primary nucleus are replicated but the MN chromatid is poorly replicated. Lollipops represent transcripts (open circles for transcripts from the normal homologue; filled circles for transcripts from the MN homologue; dashed lines for transcripts from the MN chromatid). b, Normalized transcription yield of each chromosome in a MN cell and sister cell pair after 1:3 mis-segregation. Filled and open bars indicate transcription from different homologues assessed by the parental haplotypes; filled magenta bars for the MN homologue (Chr.2B); open magenta bars for the normally segregated homologue (Chr.2A). Monosomic transcription of Chr.2B in the MN cell (bottom) is from the normally segregated chromatid in the primary nucleus and indicates near-complete silencing of the MN chromatid. c, Chromosome-wide silencing of an intact micronucleus generated by a 2:2 segregation, similar to b. The MN homologue is Chr.1B. d, Summary of transcription output in 21 MN cell–MN sister pairs, grouped by the status of MN nuclear envelope (NE) integrity. See also Supplementary Table 2 and Extended Data Fig. 3 for more information related to b–d e, RNAP2-Ser5ph signal (MN:PN ratios of background normalized fluorescent intensities) at the indicated time points after MN formation (left to right, n = 644, 212 and 605 from 2 or 3 experiments). Boxes indicate median ratio with a 95% confidence interval (CI), P values from two-tailed Mann–Whitney test. f, Correlation between micronuclei transcription (RNAP2-Ser5ph intensity) and nuclear pore complex density (POM121, 2 h after mitotic shake-off), (n = 334 from 3 experiments). Two-tailed Spearman’s correlation. g, Left, MN:PN ratios for H3K27ac (left to right, n = 187 and 118 from 2 experiments), analysed as in e. Right, correlation between H3K27ac and RNAP2-Ser5ph signals (2 h after shake-off; n = 187 from 2 experiments). Two-tailed Spearman’s correlation.