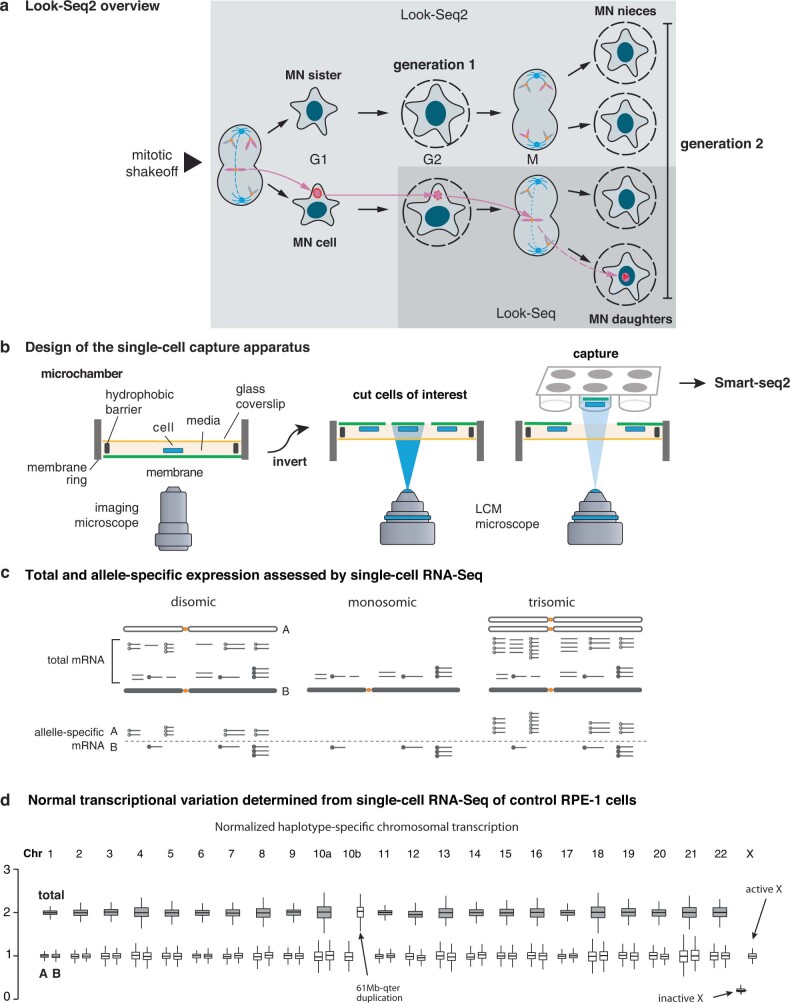
Extended Data Fig. 1. Overview of experimental and analytical workflows.
(a) Scheme of Look-Seq2. There are two key improvements compared to the original Look-Seq. First, live-cell imaging starts before the first cell division that leads to micronuclei; this enables tracking, isolation, and transcriptome analysis of both the MN cell and its sister cell (“generation 1”). Moreover, we can image cells over two cell divisions (“generation 2”) and analyze both the daughters of the MN cell (“MN daughters”) and the daughters of the MN sister cell (“MN nieces”). The second improvement is that single cells are isolated using a new capture strategy with minimal mechanical perturbation that is illustrated in (b). (b) Second generation experimental strategy for single-cell capture and sequencing. We adapted a previously developed LCM system (Palm Microbeam, Carl Zeiss) and re-designed the imaging and capture setup. The modifications enable the inversion of the membrane rings relative to the microscope objective. This allows medium to be present continuously throughout capture, which provides more time for the capture of family member cells. The setup is also compatible with laser catapulting into 96 well plates, which further increases throughput. See Methods for details. (c) Two measures of transcription yield from single-cell RNA-Seq data: (1) The total transcriptional yield is assessed by the transcripts per million (TPM) calculated from all RNA-Seq fragments overlapping with annotated coding regions. (2) The fraction of transcripts derived from each parental homologue is estimated from the counts of haplotype-specific sequencing reads. The haplotype-specific transcription yield is estimated by multiplying the total transcriptional yield by the haplotype fraction of transcripts. The transcription level of each gene in a single cell is further normalized by its mean in normal RPE-1 cells to obtain the normalized transcription of each gene. Details of the computational analysis are provided in Methods. (d) Normal range of transcriptional variation of each parental homologue derived from single-cell RNA-Seq data of control RPE-1 cells (n = 198; for Chr.12 n = 190 after excluding trisomies). Shown are the range of mean transcription of each chromosome (mean TPM ratio across all genes on a chromosome in each cell; shaded boxes) and the range of haplotype-specific transcription (mean haplotype-specific TPM ratio across all genes on a chromosome in each cell, open boxes) calculated from the total transcription and the haplotype fractions. Box plots indicate the 1st (bottom edge) and 3rd (top edge) quartiles and the median (horizontal line), with whiskers indicating 1.5x the interquartile range. The range of total transcriptional variation is used to estimate the range of normal disomic transcription (i.e., transcription of two copies of a chromosome, either from one copy of both parental homologues or from two copies of one homologue); the range of haplotype-specific transcriptional variation is used to estimate the range of normal transcription from each parental homologue. For the trisomic Chr.10q segment (61Mb-qter), the two haplotype-specific TPM ratios reflect the transcriptional output of the single-copy homologue (A) and the duplicated homologue (B); for Chr.X, the haplotype-specific TPM ratios reflect the transcriptional output of the active X (Xa) and the inactive X (Xi). For the 10q segment and Chr.X, the haplotype-specific TPM ratios are calculated by normalizing the TPM ratio of the intact 10q (A homologue) and Xa to 1. The duplicated 10q segment is appended to the q-terminus of the active X.