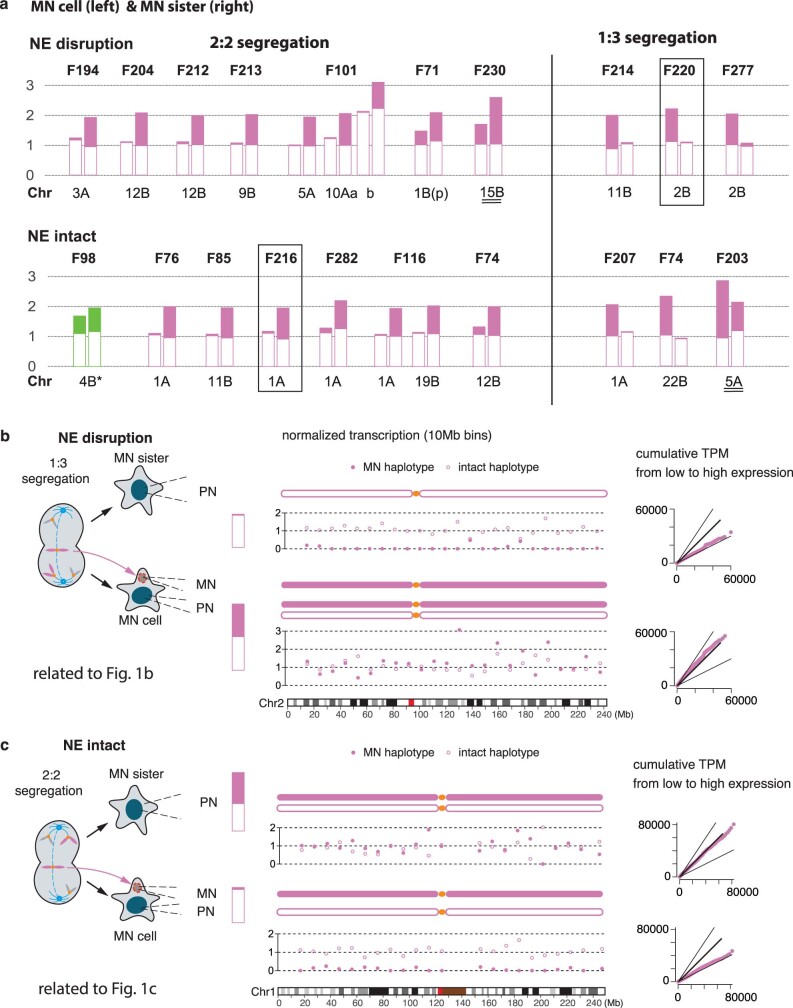
Extended Data Fig. 3. Additional data on the loss of transcription in newly generated (generation 1) MN.
(a) Summary of the transcriptional yield of MN chromosomes in all generation 1 families. Each bar plot represents the average transcriptional yield of both homologues of the MN chromosome (filled for the micronuclear homologue that is annotated below each plot; open for the intact homologue) in the MN cell (left) and the MN sister (right) in each family. Two families with near identical transcription from all chromosomes (indicating normal transcription yield of the MN chromosome) are not shown. In family F98, the MN haplotype (Chr.4B, green) displays reduced transcription in the MN cell; in all the other families, the MN haplotype in the MN cell displayed near complete silencing as indicated by either near complete loss of transcription (2:2 segregation, left) or close to monosomic transcription (1:3 segregation, right) from the intact sister chromatid of the MN haplotype in the primary nucleus. In family F71, the transcriptional imbalance is restricted to the 1p arm with near complete silencing (see Supplementary Table 2). In family F230 and F203, the transcriptional pattern indicates an extra copy of the MN homologue that is shared between the MN cell and the MN sister reflecting a pre-existing duplication of the MN homologue (i.e., a pre-existing trisomy). The examples shown in panels b (F220) and c (F216) are highlighted. (b) Chromosome-wide transcriptional data of both Chr.2 haplotypes (left) in the MN family shown in Fig. 1b (F220) and plots of cumulative TPM from low to highly expressed genes (right) plots that validate the inference of monosomic and disomic Chr.2 transcription. (c) Chromosome-wide transcriptional data of both Chr.1 haplotypes in the MN family shown in Fig. 1c (F216) and cumulative TPM plots that validate the inference of monosomic and disomic Chr.1 transcription.