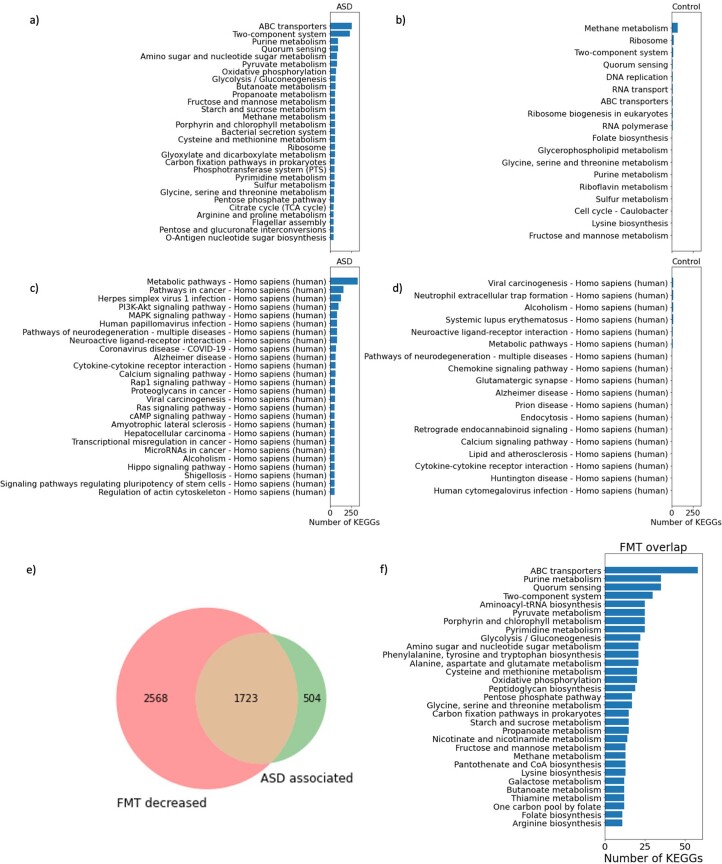
Extended Data Fig. 9. Distribution of pathways in ASD and control-associated genes detected in SMS and RNAseq data.
(a-b) Breakdown of pathways in SMS data that are associated with ASD and neurotypical controls. (c-d) Breakdown of pathways in RNAseq data that are associated with ASD and neurotypical controls. e) Overlap of ASD associated KEGG enzymes derived from the multi-cohort cross-sectional analysis and KEGG enzymes that are found to be present in the microbes that decreased in the Kang et al FMT study. f) Pathway break down of KEGG enyzmes found in both the Kang et al FMT study and ASD children in the multi-cohort cross-sectional analysis. Only microbes that were also found in the SMS data were considered in the Kang et al study.