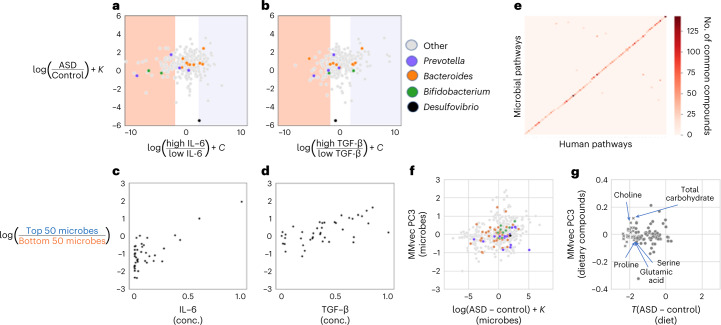
Fig. 3. Characterizing the associations among differentially abundant microbes in ASD and cytokines, gene expression in the brain and dietary patterns.
a,b, Comparison of microbial differentials obtained from age matching and sex matching and cytokine analysis. c,d, Microbial log ratios constructed from the 50 top and bottom most differentially abundant microbes corresponding to each cytokine. K and C represent unknown biases due to the shift in the microbial load between the ASD and neurotypical control population. e, Heat map showing the overlap of molecules between ASD-enriched pathways in the microbiome and in the brain. The microbial and human pathways are both sorted alpha-numerically; the dense diagonal is largely indicative of common pathways between microbial and human genomes. f,g, PC3 from microbe–diet co-occurrence analysis is contrasted against microbial log fold changes and dietary differences from Berding et al.25 Dietary compounds that are depleted (P < 0.1) in children with ASD are highlighted as ‘x’ markers. T(ASD-Control) represents the t-statistic that measures the differences between ASD and neurotypical dietary intake. conc., concentration.