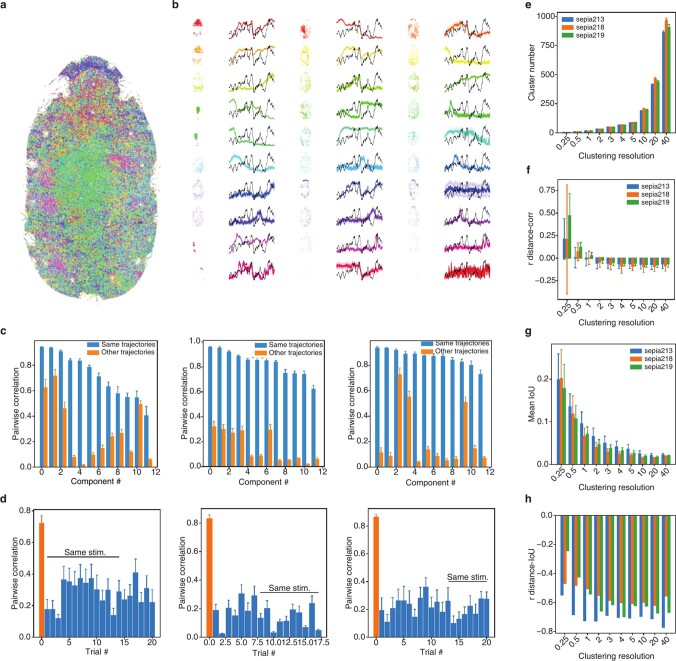
Extended Data Fig. 6. Chromatophore component clustering.
a. Clustering of chromatophores based on the activity in the trajectory in Fig. 4a (Leiden community detection, N = 30 clusters). b. Spatial distribution of chromatophores in each cluster (insets at left) and their average area change over time in this transition. Black trace common to all panels is the animal’s speed of pattern change. c. Averaged pairwise correlation of chromatophore activity within pattern components. The top 12 most variable components are plotted. Blue, correlation of chromatophore activity during the trajectory in which pattern components are defined. Orange, average correlation of activity of the same chromatophores during other trajectories (N = 21, 18, 21 trials for sepia213, left; sepia218, middle; sepia219, right). d. Correlation as in c, averaged over components 1-12 instead of trials. Orange: average correlation in the selected camouflage transition. Blue: correlation of same clustering during other trajectories. (grouped by stimulation types; N = 21, 18, 21 trials for sepia213, left; sepia218, middle; sepia219, right). e. Number of clusters at different clustering resolutions. Clustering was based on trial-specific activity. (N = 21, 18, 21 trials for sepia213, sepia218, sepia219). f. Absence of correlation between Wasserstein distance and group activity correlation (Mean ± s.e.m. of Pearson’s r; as in Fig. 4d) at different clustering resolutions. g. Mean IoU (Intersection over Union of chromatophore groupings) at different clustering resolutions (Mean ± s.e.m.). Higher clustering resolution does not result in higher clustering stability. h. Correlation coefficient of the distance between trajectories and mean IoU (Mean ± s.e.m. of Pearson’s r; as in Fig. 4i), at different clustering resolutions.