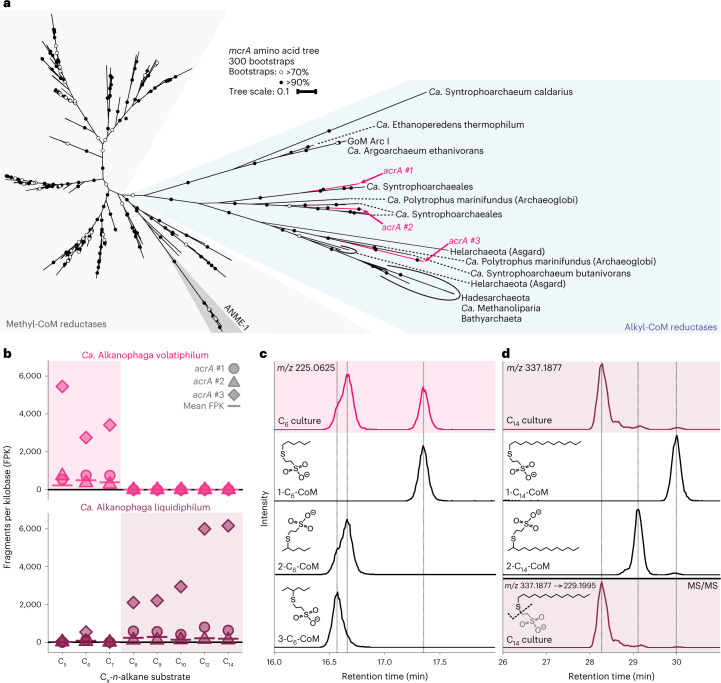
Fig. 3. Ca. Alkanophaga use alkyl-coenzyme M reductases to activate alkanes to alkyl-CoMs.
a, Phylogenetic placement of translated mcrA sequences of Ca. Alkanophaga. Both Ca. Alkanophaga species contain three mcrA sequences, all of which fall into the divergent branch of mcrAs, encoding alkyl-CoM reductases (Acrs), highlighted in blue. The six acrA sequences form three clusters of two sequences, each cluster containing one sequence of each Ca. Alkanophaga species. Tree scale bar, 10% sequence divergence. b, Expression of acrA genes during growth on various alkanes for both Ca. Alkanophaga species. Cultures in which the respective species was prevalent in the metagenomes are highlighted with shaded boxes. The mean expression of all genes of the respective species is shown as a horizontal bar. The acrA of the third cluster was strongly expressed, irrespective of substrate length, by the species abundant in that culture. The expression of the other acrA genes was low. c,d, Extracted ion chromatograms (EICs) based on exact mass and a window of ±10 mDa of deprotonated ions of variants of C6-CoM (c) and C14-CoM (d) detected via liquid chromatography–mass spectrometry. In both c and d, the upper shaded panels show the culture extract, with isomers of alkyl-CoM standards below. In d, the shaded bottom panel shows the EIC produced with the exact mass of the C14-thiolate, a fragmentation product derived in MS/MS experiments from the precursor C14-CoM. Dashed vertical lines were added at retention times of peak maxima of standards (c) or standards and fragmentation products (d) for easier identification of peaks in the culture extracts. While C6 is activated on the first and second carbon atom to a similar degree, C14 is activated predominantly to ≥3-C14-CoM.