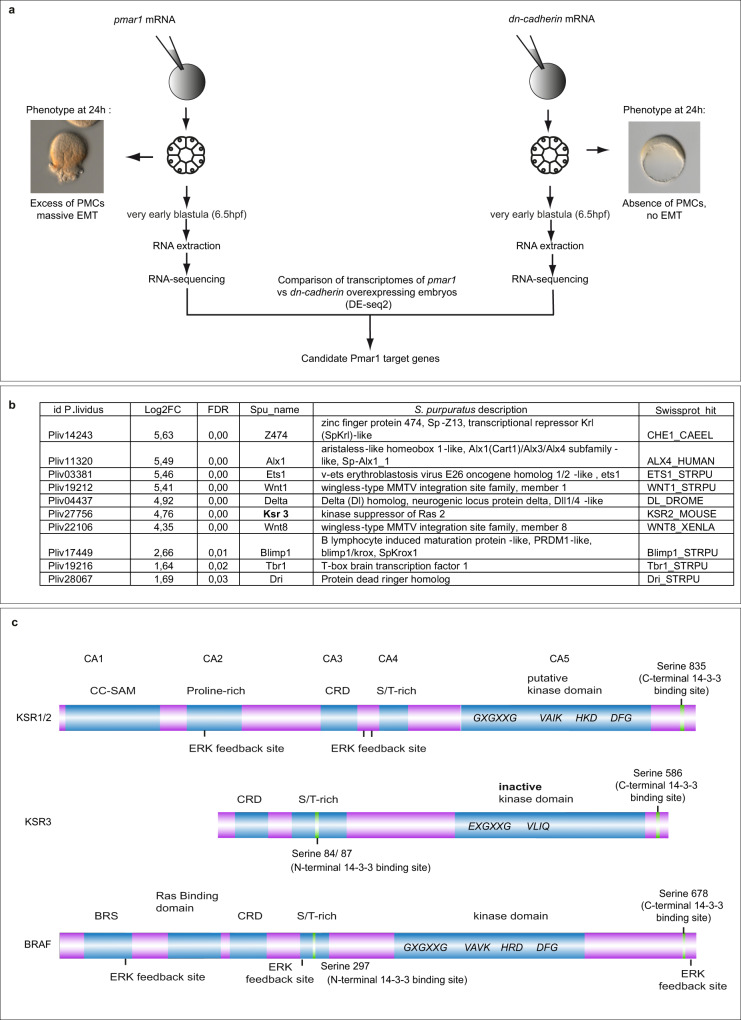
Fig. 2. Transcriptome profiling identifies Kinase Suppressor of Ras 3 as a candidate for activation of ERK in the skeletogenic precursors.
a Design of the screen. pmar1 overexpression induces a massive activation of ERK and causes formation of an excess of PMCs while overexpression of DN-cadherin prevents expression of pmar1, blocks activation of ERK, suppresses formation of PMCs and eliminates expression of vegetal pole marker genes by sequestering β-catenin at the level of the membrane. RNA profiling of embryos overexpressing pmar1 or dn-cadherin should therefore allow to identify the gene(s) that activates ERK signalling. b Top list of positive controls and novel genes recovered in the screen for genes upregulated following overexpression of pmar1. The identification numbers of the Paracentrotus lividus gene predictions are given in the first column, together with the Log2 (fold change) between the pmar1 overexpression versus dn-cadherin overexpression conditions. FDR, false discovery rate. Spu, Strongylocentrotus purpuratus. All the genes involved in activation of the PMC GRN in different species were present in the list of candidates genes upregulated in pmar1 overexpressing embryos compared to DN-cadherin overexpressing embryos. The only gene related to MAPK/ERK signaling in this list of candidates encoded a member of the Kinase Suppressor of Ras family: KSR3. c Domain organization of the sea urchin KSR1, KSR3 and BRAF. KSRs and RAF proteins share conserved domains such as the kinase domain, the S/T rich domain and the cysteine rich domain or CRD (also called C1 domain). In contrast the BRAF-specific domain (BRS) and the RAS binding domain (RBD) are uniquely present in BRAF while the CC-SAM domain is present in the sea urchin KSR1 but absent in KSR3. Several motifs highly conserved in kinases are also missing or highly divergent in KSR3 including the GXGXXG and VAV/IK motif of the ATP binding site, the HRD motif of the catalytic loop, and the DFG motif of the activation loop.