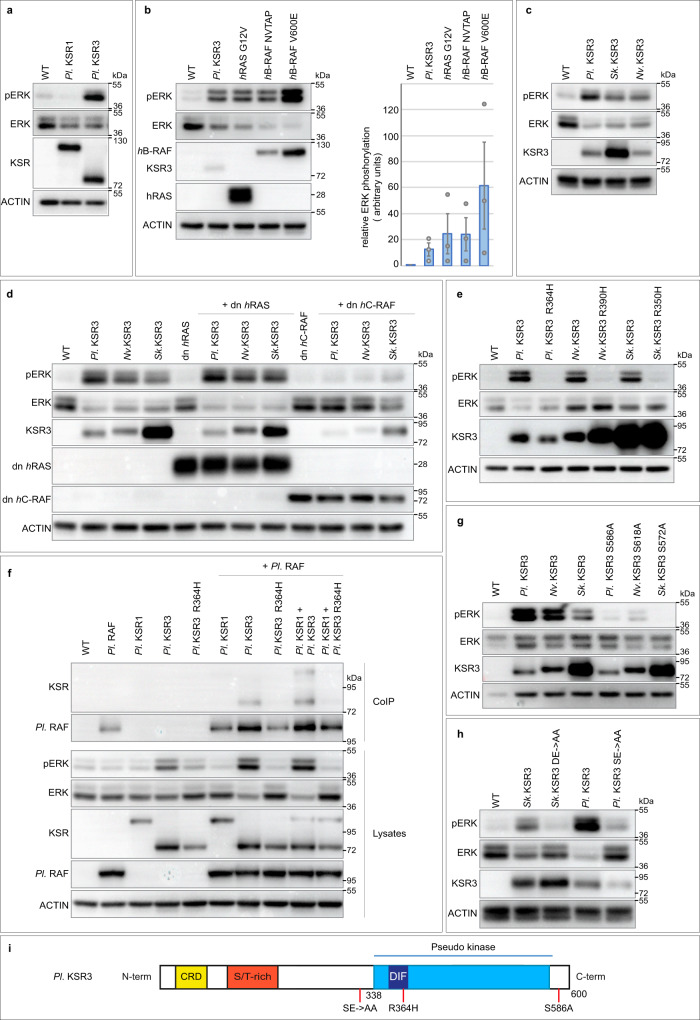
Fig. 6. KSR3 factors act as RAS-independent constitutively active triggers of ERK signaling.
a Overexpression of sea urchin KSR3 (Pl. KSR3) but not of KSR1 (Pl. KSR1) triggers ERK signalling in Human HEK293T cells. b Overexpression of sea urchin KSR3 activates ERK signalling at the same level as overexpression of activated RAS G12V or of an activated B-RAF deletion mutant (Delta NVTAP). c KSR3 from Saccoglossus kovaleskii (Sk. KSR3) and from Nematostella vectensis (Nv. KSR3) also activate ERK signalling when overexpressed. d KSR3 from sea urchin, hemichordate and cnidarian activate ERK signalling in a RAS-independent but RAF-dependent manner. e KSR3 from echinoderm, hemichordate and cnidarian require dimerization to activate ERK signalling. Substitution of a highly conserved arginine residue required for dimerization by histidine suppressed the ability of KSR3 proteins to activate ERK signalling. f Sea urchin KSR3 (Pl KSR3) interacts with sea urchin RAF in co-immunoprecipitation experiments. g KSR3 factors from Paracentrotus, Saccoglossus and Nematostella require interaction with 14-3-3 proteins at the C-terminus. Substitution of alanine for a serine residue located in a potential 14-3-3 binding site of sea urchin (sequence of the motif: HSLSEP), hemichordate (HSQSEP) or cnidarian (GSHSEP) strongly impairs the ability of these factors to activate ERK signalling. h Phosphorylation of the NtA motif of KSR3 factors is essential for their ability to activate ERK signalling. Substitution of alanine for the aspartate and glutamate residues 293, 294 of Saccoglossus KSR3 and for serine 306 and glutamate 307 of Paracentrotus KSR3 strongly impairs their ability to activate ERK signalling. i Scheme describing the different mutations analysed. Source data are provided as a Source Data file. Information about quantification and replication of the western blots are provided in Supplementary Table 1.