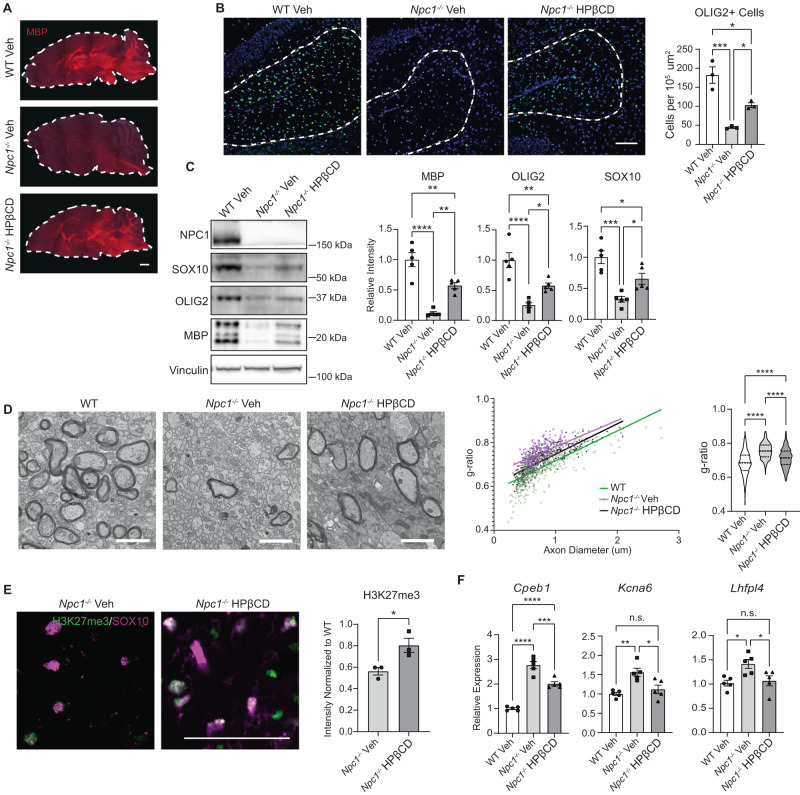
Fig. 7. A single injection of 2-hydroxypropyl-β-cyclodextrin rescues myelination defects.
Mice were administered 2-hydroxypropyl-β-cyclodextrin (4000 mg/kg, i.p.) (HPβCD) or a vehicle control (saline) (Veh) at P7. Brains were collected and myelination assessed at P16. Dashed line in A indicates the outline of the brain section. Midline sagittal sections were stained for (A) MBP (red) and (B) OLIG2 (green). Nuclei stained with DAPI (blue). The number of OLIG2+ cells in the corpus callosum (dashed line in B) quantified at right. C Western blot for SOX10, OLIG2, and MBP from forebrain lysates. Values were normalized to vinculin and relative to WT. Quantified at right. D TEM images of the corpus callosum. G-ratios were quantified (right). E Midline sagittal sections were stained for H3K27me3 (green) and SOX10 (magenta). Images were captured from the corpus callosum, and the intensity of H3K27me3 within SOX10+ nuclei was quantified (right). F Expression of neuronal genes in O4+ cells purified from P16 mice as determined by qRT-PCR. A Scale bars = 1000 µm; (B) 100 µm; (D) 2 µm; (E) 100 µm. A Experiment was performed on 3 biological replicates per group, all with similar results; dashed lines outline the sagittal brain. D Dashed lines in the violin plot indicate quartiles. Data are mean ± SEM. n.s. not significant, *p < 0.05, **p < 0.005, ***p < 0.001, ****p < 0.0001 by one-way ANOVA with Tukey post hoc test and multiple comparisons (B–D, F) or two-tailed unpaired Student’s t-test (E). B n = 3 mice; F = 27.65; df = 2; p = 0.0009 (C) n = 5 mice; F = 35.97, 22.49, 16.55; df = 2; p = <0.0001, <0.0001, 0.0004 (D) Linear regression slopes: F = 1.574; df = 2; p = 0.2078; Linear regression intercepts: F = 256.9; df = 2; p < 0.0001; ANOVA: n = 443, 301, 327 axons (from 3 mice per group); F = 128.1; df = 2; p = <0.0001 (E) n = 3 mice; t = 3.167; df = 4; p = 0.0340 (F) n = 5 mice; F = 78.24, 10.83, 6.170; df = 2; p = <0.0001, 0.0021, 0.0144. Source data are provided as a Source Data file.