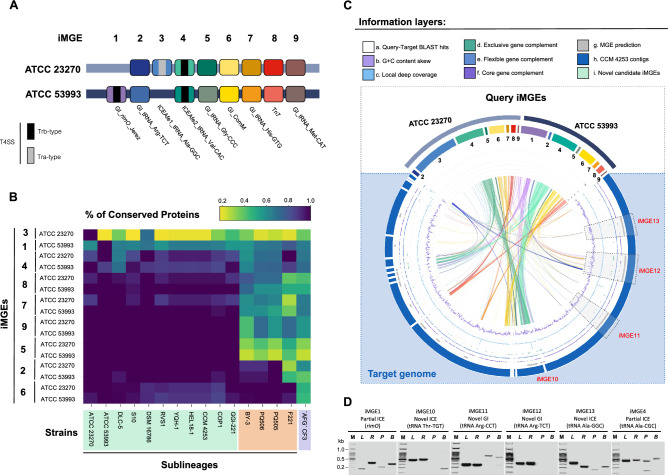
Figure 3.
Location, occurrence, and mapping of MGE-associated genes from validated and candidate iMGEs in the draft genome of A. ferrooxidans CCM 4253. (A) Location of experimentally validated and bioinformatically predicted iMGEs (candidate) present in the two query genomes and their correlative order (iMGE1 to iMGE9). Query iMGEs were classified as Tra- or Trb-type Integrative Conjugative Elements (ICEs), Genomic Islands (GIs), or Tn7 transposons (Tnp) and their integration sites were identified (detailed in Table 1). (B) Occurrence and coverage of validated and candidate iMGEs of A. ferrooxidans ATCC 23270T and ATCC 53993 in an extended set of draft genomes of the species. Strains were clustered based on the coverage patterns of the MGE-associated gene products (cut-off: e-value -10; detailed in Supplementary Table 4). (C) Identification of novel iMGEs and iMGE-fragments in strain CCM 4253. Colored elements in the outer layer are known and candidate iMGEs in ATCC 23270T and ATCC 53993 genomes. Some iMGEs are strain-exclusive, such as iMGE120 and iMGE323. Others are conserved in two reference strains used as queries. Dark blue elements correspond to CCM 4253 strain genomic assembly contigs (GCA_003233765). Additional information layers, from the center of the figure outwards, correspond to (a) the TBlastN hits found in the CCM 4253 genome using either query strains MGEs; (b) the G + C content skew of the position; (c) the deep local coverage at the position; (d–f) the exclusive, flexible, or core pangenome compartment to which a given CCM 4253 gene pertains and (g) the prediction of MGE features using a combination of tools as previously described in Gonzalez et al.40 and Moya-Beltrán et al.33. (D) PCR validation of novel and partially conserved candidate iMGEs identified in the genome of strain CCM 4253. PCR products correspond to specific attL, attR, attP, and attB sites (the scheme of experimental design is shown in Supplementary Table 4D and primer sequences are listed in Supplementary Table 4E) of the following iMGEs: iMGE1 (partial ICE integrated into the rimO gene); iMGE10 (integrated at tRNA Thr-TGT); iMGE11 (integrated at tRNA Arg-CCT); iMGE12 (integrated at tRNA Arg-TCT); iMGE13 (integrated at tRNA Ala-GGC); iMGE4 (partial ICE integrated at tRNA Ala-CGC). PCR of attP and attB sites were evaluated on DNA recovered after incubation of A. ferrooxidans CCM 4253 cells with mitomycin C. Lane M represents the 100 bp DNA ladder; lanes L, R, P, and B represent the attL, attR, attP, and attB specific sites in the individual iMGEs, respectively. The gel image has been cropped for display. The original gel image is shown in Supplementary Table 4F.