FIGURE 1.
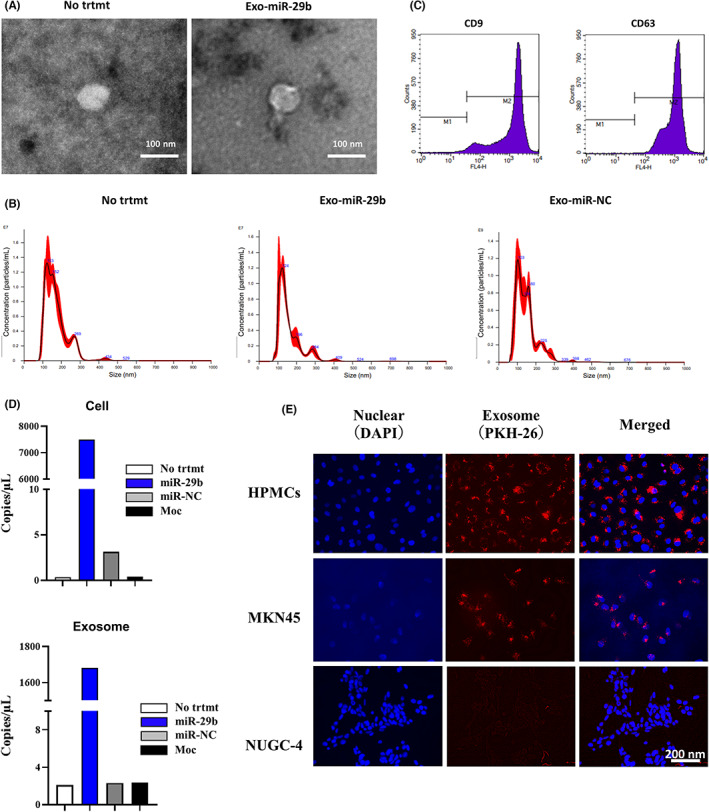
Identification of extracellular vesicles (EVs) derived from human bone marrow‐derived mesenchymal stem cells (BMSCs). (A) EVs isolated from supernatant of human BMSCs (UE6E7T‐12) (No treatment [trtmt]), BMSCs transfected with microRNA (miR)‐29b (Exo‐miR‐29b), or negative control (Exo‐miR‐NC) were dropped onto a copper grid, dyed with uranyl acetate, and observed using an HT‐7700 transmission electron microscope. Magnification, 40,000×. (B) Identification of EVs by nanoparticle tracking analysis. EV samples were observed for size distribution and number of extracellular vesicles by NanoSight LM10. (C) Detection of EV surface antigens by flow cytometry. Latex beads were incubated with EV and EV‐coated beads were analyzed for the presence of mAbs to CD9 or CD63 by a flow cytometer. (D) miR‐29b expression in human BMSCs (UE6E7T‐12) and corresponding EVs were quantified with 3D‐digital PCR. UE6E7T‐12 was transfected with recombinant lentiviral vector containing miR‐29b or miR‐negative control (miR‐NC) vector or vector alone (Moc) with 10 μg/mL polyacrylamide. After 20 h, the medium was replaced with fresh culture medium. Three days after transfection, EVs were extracted from the UE6E7T‐12 supernatant by ultracentrifugation. miRNAs were isolated from cell pellets and EVs using the miRNeasy kit. Experiments were performed in singleplex by the QuantStudio 3D Digital PCR System platform. (E) EVs derived from UE6E7T‐12 were stained with PKH26 and cocultured with HPMCs, MKN‐45, or NUGC‐4 cells for 24 h and observed with fluorescein microscopy. Magnification, 400×.
