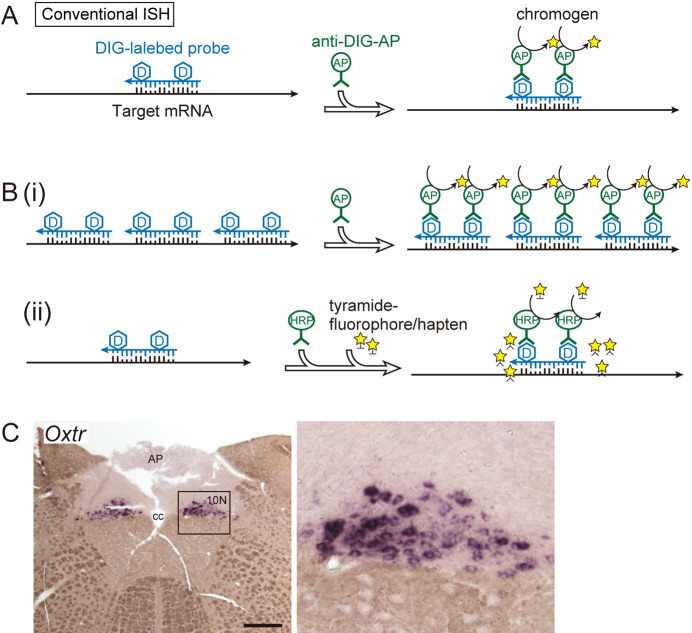
Fig. 1.
Schematic of conventional in situ hybridization. A: DIG-labeled in situ hybridization. RNA probes complementary to target sequences were used. A chromogenic reaction using an alkaline phosphatase-labeled antibody is shown as an example of a signal detection method. B: Examples of signal amplification methods for conventional in situ hybridization. (i) Using multiple probes to increase coverage of the target mRNA sequence. The signal strength is proportional to the total probe length. (ii) Combination with TSA amplification. Tyramide activated by peroxidase is anchored to tyrosine residues of the surrounding protein. C: Example of visualization of low-expression genes in the rat brain. Oxtr, which encodes the oxytocin receptor, is visualized in the vagus nerve nucleus (10N) of the medulla oblongata by conventional in situ hybridization without TSA amplification. The right panel is a magnified image of the framed area in the left micrograph. Bar = 500 μm. AP, area postrema; cc, central canal.