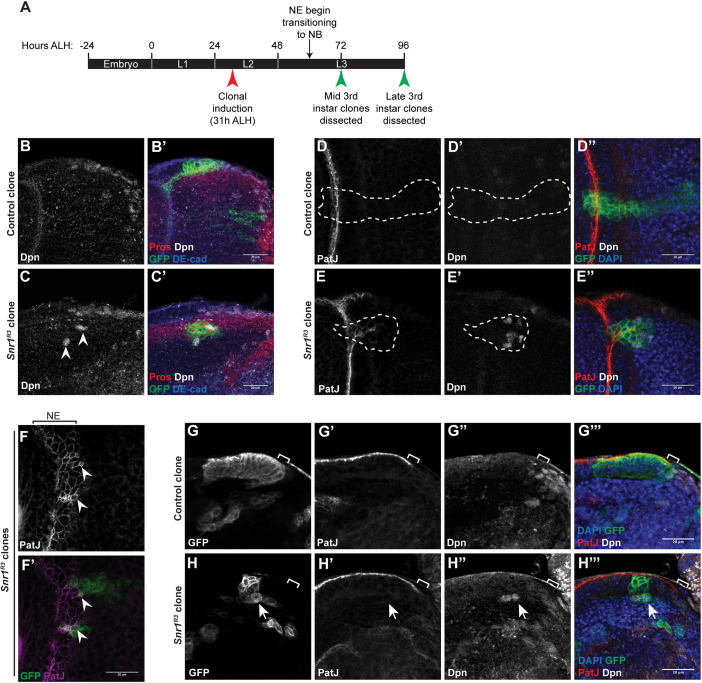
Fig. 2.
Snr1 is required for the transition from neuroepithelial cells to neuroblasts. (A) Timeline of larval development showing timing of clonal induction and dissections after larval hatching (ALH). (B-C′) Snr1 mutant clones in the optic lobe have altered location and morphology. (B,B′) Control MARCM clone at surface of brain imaged in cross-section. (C,C′) Snr1R3 MARCM clone located deeper inside the brain showing ectopic expression of Deadpan (Dpn; arrowheads). Cell junctions are marked by DE-cad, neuroblasts are marked by Dpn, MARCM clones are marked by GFP and GMCs are marked by Pros. (D-E″) Snr1 mutant clones deep in the medulla express markers for neuroepithelial cells and neuroblasts. (D-D″) Control optic lobe MARCM clone marked by GFP. Clone is outlined by a white dashed line. (E-E″) Snr1R3 optic lobe MARCM clone marked by GFP. Clone is outlined by a white dashed line. Neuroepithelial cells marked by PatJ (red) and neuroblasts marked by Dpn (white). (F-F′) Snr1R3 clone cells on the surface of the brain are smaller (arrowheads) and express neuroepithelial (NE) cell marker. Neuroepithelial cell junctions are marked by PatJ (magenta) and MARCM clones are marked by GFP (green). (G-H‴) MARCM clones imaged in cross-section at mid third instar stage. Brackets indicate transition zone. Arrows indicate ectopic Dpn+ cells. Scale bars: 20 µm.