Abstract
Coronavirus disease 2019 (COVID-19) is an acute respiratory infection caused by the severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2). The uncontrolled systemic inflammatory response, resulting from the release of large amounts of pro-inflammatory cytokines, is the main mechanism behind severe acute respiratory syndrome and multiple organ failure, the two main causes of death in COVID-19. Epigenetic mechanisms, such as gene expression regulation by microRNAs (miRs), may be at the basis of the immunological changes associated with COVID-19. Therefore, the main objective of the study was to evaluate whether the expression of miRNAs upon hospital admission could predict the risk of fatal COVID-19. To evaluate the level of circulating miRNAs, we used serum samples of COVID-19 patients collected upon hospital admission. Screening of differentially expressed miRNAs in fatal COVID-19 was performed by miRNA-Seq and the validation of miRNAs by reverse transcription-quantitative polymerase chain reaction (RT-qPCR). The Mann–Whitney test and receiver operating characteristic (ROC) curve were used to validate the miRNAs, whose potential signaling pathways and biological processes were identified through an in silico approach. A cohort of 100 COVID-19 patients was included in this study. By comparing the circulating levels of miRs between survivors and patients who died due to complications of the infection, we found that the expression of miR-205-5p was increased in those who died during hospitalization, and the expression of both miR-205-5p (area under the curve [AUC] = 0.62, 95% confidence interval [CI] = 0.5–0.7, P = 0.03) and miR-206 (AUC = 0.62, 95% CI = 0.5–0.7, P = 0.03) was increased in those who lately evolved to severe forms of the disease (AUC = 0.70, 95% CI = 0.6–0.8, P = 0.002).“In silico” analysis revealed that miR-205-5p has the potential to enhance the activation of NLPR3 inflammasome and to inhibit vascular endothelial growth factor (VEGF) pathways. Impaired innate immune response against SARS-CoV-2 may be explained by epigenetic mechanisms, which could form early biomarkers of adverse outcomes.
Keywords: COVID-19, SARS-CoV-2, microRNAs
Impact statement
Even after the advent of vaccination, severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2) still circulates and causes death worldwide. Epigenetic mechanisms, such as gene expression regulation by microRNAs, may be at the basis of the individual immunological responses associated with coronavirus disease 2019 (COVID-19), and also related to the increased risk of poor outcomes in some people. In this way, the identification of new serum prognostic biomarkers at the time of the hospitalization is a feasible approach to improving patient care, and also filling gaps in the knowledge of the molecular pathways of the disease that are still not completely understood.
Introduction
Since 2019, the novel coronavirus disease 2019 (COVID-19) has spread worldwide leading to global health instability and millions of deaths.1,2 Even after the advent of vaccines, new COVID-19 fatal cases are still being reported. 3 Clinical manifestations of COVID-19 range from mild flu-like symptoms to multiple organ failure and death, and the prediction of fatal cases remains challenging.4,5 These differences in the individual responses to the disease have been attributed to genetic and epigenetic variations.3,6
MiRs consist of small, non-coding, single-stranded RNA molecules that play an important role in the post-transcriptional regulation of gene expression mainly by direct binding to messenger RNAs (mRNAs).7,8 Furthermore, miRs are actively secreted into the bloodstream, participating in intercellular communication between several cells and tissues and possibly orchestrating pathological processes by mechanisms of endocrine-genetic signaling. 9
In the last decade, miRNAs have been associated with several diseases, including infections such as HIV 10 and hepatitis C. 11 However, despite miRs have been pointed as epigenetic modulators of immune response against COVID-1912,13 and other diseases caused by coronaviruses, 14 the association between circulating miRs and the risk of death in COVID-19 remains to be explored. Therefore, the aim of this study was to evaluate whether the expression of specific miRs upon hospital admission could predict the risk of fatal COVID-19.
Materials and methods
Selection and clinical features of COVID-19 patients
This study cohort comprised adult patients with confirmed diagnosis of COVID-19, admitted to the UNICAMP Clinical Hospital, one of the largest public hospitals in South America, due to COVID-19 manifestations. Patients were enrolled from January to March 2021, during the second wave of the COVID-19 in Brazil. Patients of both genders aged between 18 and 98 years were eligible to the study. The diagnosis of COVID-19 was confirmed by real-time polymerase chain reaction (RT-PCR). Patients with two negative severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2) RT-PCR results and no radiological findings were considered COVID-19 negative and were excluded from the study. We also excluded patients with cancer; organ transplantation; systemic chronic infections; pregnant women; neurodegenerative, autoimmune, chronic kidney, or hepatic diseases; or patients within less than 24 h of hospitalization.
COVID-19 patients’ serum samples were obtained from the clinical laboratory of UNICAMP Clinical Hospital, to where the patients’ blood samples were sent for routine analysis. Only the blood samples collected on the first day of hospitalization were considered for this study.
Patients were followed up from the day of admission until the day of discharge or death. All demographic and clinical data were daily retrieved from electronic medical records during the hospitalization period. Two outcomes were recorded: critical COVID-19 and death during hospitalization.
Isolation and identification of miRs differentially expressed in fatal COVID-19 by next-generation sequencing
First, we extracted total RNA from the serum samples using the miRNeasy Serum/Plasma Kit (Cat. No. / ID: 217184; Qiagen), and by next-generation sequencing (NGS) approach, we screened dysregulated miRNAs in fatal COVID-19 compared to survivors. MiRs Library kit (Cat. No. / ID: 331502; Qiagen) was used to obtain complementary DNA (cDNA) libraries, which were sequenced in a MiSeq sequencer using the Reagent Kit v3, 150-cycle (MS-102-3001; Illumina). A total of 2,000,000–5,000,000 reads per sample were obtained. All procedures were performed according to the manufacturers’ recommendations and the library quality control accessed by concentration (Qubit Fluorometric Quantitation; Thermo Fisher Scientific) and size (2100 Bioanalyzer Instrument; Agilent).
NGS differential expression data analysis between fatal COVID-19 and survivors
Data generated by NGS were aligned against the reference genome and normalized using the DESeq2 method in GeneGlobe Data Analysis Center (Qiagen) software. Next, we performed differential expression analyses between the two groups (fatal COVID-19 and survivors) according to the unique molecular index (UMI) count in the samples. We considered differentially expressed miRs with fold change ±2 and a value of P < 0.01. In addition, we also performed the differential expression analyses as previously described 14 using R script analysis (R CoreTeam, 2017) and we considered adjusted P value (adj. P) <0.05 to detect differentially expressed miRs between the two groups. Finally, we considered miRs differentially expressed between fatal COVID-19 and survivors, only those miRs that showed differential expression in both methods.
In silico prediction of miR targets and biological function validation
Differentially expressed miR predicted target genes were accessed using Ingenuity Pathway Analysis (IPA®, Qiagen bioinformatics) and miRWalk 3.0 software. Gene set enrichment analysis was submitted to DAVID annotation (https://david.ncifcrf.gov/) tool, and enriched pathways were considered when the P value was <0.05.
Validation of differentially expressed miRs in an expanded cohort by quantitative polymerase chain reaction
RNA extracted from COVID-19 patients’ serum samples (miRNeasy Serum/Plasma Kit, Cat. No. / ID: 217184; Qiagen) was reversely transcribed into cDNA using miRCURY LNA RT kit (Cat. No. / ID: 339340; Qiagen). Quantitative polymerase chain reaction (qPCR) expression was quantified using the ABI 7500 Sequence Detection System (Applied Biosystems) after SYBR green master mix addition (miRCURY LNA SYBR Green PCR Kit, Cat. No. / ID: 339306; Qiagen). We used the Pfaffl method 15 to calculate the relative miRNA expression, using endogenous hsa-miR-16-5p (GeneGlobe ID – YP00205702) as a reference gene and exogenous cel-miR-39 (GeneGlobe ID – YP00203952) as an interplate calibrator. All reactions were performed in duplicates and primers were validated by the supplier in wet lab with estimated PCR efficiency of 2 (miRCURY LNA miRNA PCR Assay, Cat. No. / ID: 339346, hsa-miR-133a-3p – GeneGlobe ID – YP00204788, hsa-miR-206 – GeneGlobe ID – YP00206073, hsa-miR-205-5p – GeneGlobe ID – YP00204487, hsa-miR-22-3p – GeneGlobe ID – YP00204606, hsa-miR-4516 – GeneGlobe ID – YP02112882, hsa-miR-184 – GeneGlobe ID – YP00204601; Qiagen). Negative controls were included on each run and produced no amplification.
Statistical analysis
We performed descriptive analyses to describe patients’ baseline characteristics and clinical outcomes. Categorical variables were expressed in absolute number and percentage. Numerical variables with normal distribution were expressed as mean and standard deviation, and numerical variables with non-normal distribution were expressed as median and interquartile range (IQR). MiR expression levels were compared between severe and non-severe COVID-19, and fatal COVID-19 and survivors using the Mann–Whitney test. Their discriminatory capacity to distinguish between the two groups was accessed by receiver operating characteristic (ROC) analyses. GraphPad Prism version 6.0 (GraphPad Software Inc., La Jolla, CA, USA) was used for graph plotting, and SPSS version 25.0 for Windows (SPSS Inc., Chicago, IL, USA) was used for statistical analysis.
Results
Screening cohort
The screening cohort for the analysis of the circulating differentially expressed miRs was composed of 10 survivors and 7 fatal COVID-19 patients. In both groups, most of the patients were middle-aged men (survivors’ median age = 59 years, fatal COVID-19 median age = 55 years) with history of a previous disease, such as hypertension, diabetes, or obesity. Clinical and demographic features of the screening cohort were described in Table 1.
Table 1.
Demographic and clinical data of COVID-19 patients selected for the screening cohort.
| Clinical and demographic features | Survivors (n = 10) | Fatal COVID-19 (n = 7) |
|---|---|---|
| Age in years, median (IQR) | 59 (50.2–66.2) | 55 (45.0–65.0) |
| Male, n (%) | 6 (60.0) | 5 (71.4) |
| Previous diseases, n (%) | ||
| Hypertension | 6 (60.0) | 3 (42.9) |
| Diabetes | 2 (20.0) | 2 (28.6) |
| Obesity | 3 (30.0) | 3 (42.9) |
IQR: interquartile range; n: absolute number; COVID-19: coronavirus disease 2019.
miR differential expression in the screening cohort patients
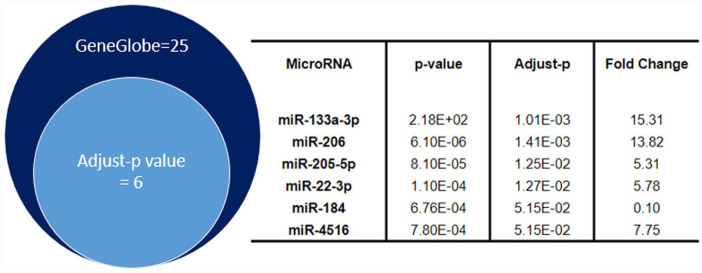
By an NGS analysis approach, we found a total of 25 miRs differentially expressed between survivors and fatal COVID-19 patients when considering P < 0.01 in GeneGlobe analysis (data not showed). Among them, only miR-133a-3p, miR-206, miR-205-5p, miR-22-3p, miR-184, and miR-4516 remained dysregulated when the adj. P of the sample UMI count (adj. P < 0.05) was considered. These results are illustrated in Figure 1, in which the top-six miRNAs differentially expressed in fatal COVID-19 were ranked according to the adj. P.
Figure 1.
miRs differentially expressed in survivors and fatal COVID-19 detected by NGS in COVID-19 patients.
miRs: microRNAs; COVID-19: coronavirus disease 2019; NGS: next-generation sequencing.
Using GeneGlobe analysis, a total of 25 dysregulated miRNAs were detected in fatal (P < 0.01) COVID-19 patients compared to the survivors. This result was graphically represented by the dark-blue circle on the left side. Six of these 25 miRs, miR-133a-3p, miR-206, has-miR-205-5p, miR-22-3p, miR-184, and miR4516 (pale-blue circle on the left side) remained differently expressed between the two groups (adj. P < 0.05) when we performed the adjustment of the P value of the expression of miRs in R script analyses. Statistical characterization of these six miRNAs is described on the right side of this figure.
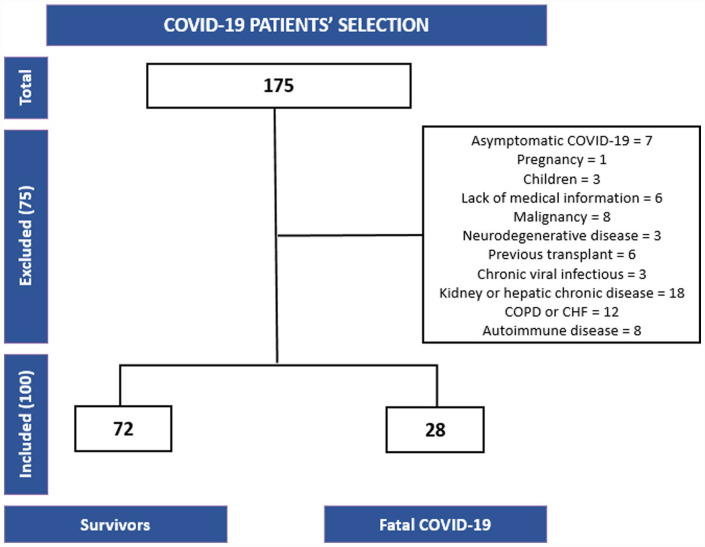
Patient inclusion and exclusion criteria
A total of 100 hospitalized patients due to COVID-19-related respiratory symptoms composed our validation cohort. The inclusion and exclusion criteria of the patients are detailed in Figure 2.
Figure 2.
Flowchart of COVID-19 patient selection, inclusion, and reasons for exclusion.
COPD: chronic obstructive pulmonary disease; CHF: chronic heart failure; COVID-19: coronavirus disease 2019.
Patient clinical and demographic features
The validation cohort was composed of 100 hospitalized patients due to COVID-19. Among those, 28 patients died due to COVID-19 complications in the hospital, 27 were critical COVID-19 survivors, and 45 patients had non-critical COVID-19. Patient demographic and clinical features were described according to their outcome (non-critical patients, critical COVID-19 survivors, or fatal COVID-19) in Table 2. Non-survivors were slightly older than critical COVID-19 survivors and non-critical COVID-19 (median age: 61.5 years versus 57.0 and 54.0 years, respectively). Also, individuals with hypertension (64.3%), diabetes (46.4%), obesity (24.8%), and cardiovascular diseases (25.0%) were more prevalent among the non-survivors group, but this difference was not observed between critical COVID-19 survivors and non-critical patients.
Table 2.
Demographic and clinical data of the entire COVID-19 cohort.
| Clinical and demographic features | Non-critical survivors (n = 45) | Critical survivors (n = 27) | Fatal COVID-19 (n = 28) |
|---|---|---|---|
| Age in years, median (IQR) | 57.0 (42.0–66.0) | 54.0 (44.0–63.0) | 61.5 (50.2–71.3) |
| Male, n (%) | 21 (46.6) | 16 (59.2) | 18 (64.3) |
| Previous diseases, n (%) | |||
| Hypertension | 16 (35.5) | 10 (37.0) | 18 (64.3) |
| Diabetes | 13 (28.8) | 7 (29.9) | 13 (46.4) |
| Obesity | 10 (22.2) | 10 (37.3) | 12 (24.8) |
| Cardiovascular diseases a | 4 (8.8) | 3 (11.1) | 7 (25.0) |
IQR: interquartile range; n: absolute number; COVID-19: coronavirus disease 2019.
Cardiovascular diseases = previous heart attack, arrhythmia, and heart failure.
Association between differentially expressed miRs and critical COVID-19
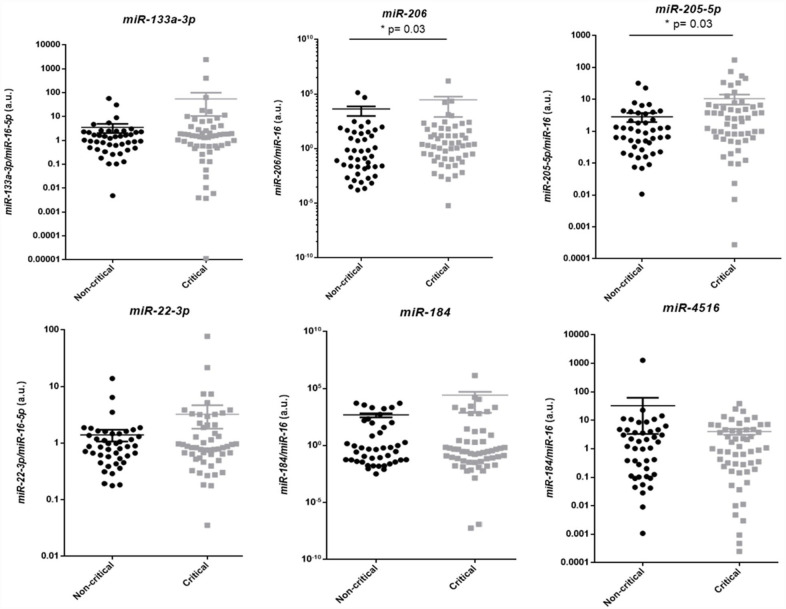
Serum levels of the top-six miRs identified by NGS analysis were validated in the validation cohort of COVID-19 patients by qPCR. To evaluate if the top-six miRs were related to an increased risk of the development of critical COVID-19 forms, we first compared the miR expression levels in serum of patients who needed intensive care and/or died at any time of the hospitalization period (critical survivors and fatal COVID-19, n = 55) with the serum levels of patients who were treated on the ward and survived (non-critical survivors, n = 45).
As a result, we found that both miR-205-5p (P = 0.03) and miR-206 (P = 0.03) were upregulated at the beginning of the hospitalization in patients who presented critical COVID-19 compared to non-critical ones (Figure 3).
Figure 3.
miR validation in the expanded cohort of COVID-19 patients, comparing critical versus non-critical patients.
miR: microRNA; COVID-19: coronavirus disease 2019; IQR: interquartile range.
MiR-133a-3p (non-critical: n = 45, median [M] = 1.1, IQR 35 = 0.5–2.3; critical COVID-19: n = 55, M = 0.5, IQR = 1.5–3.5; P = 0.5). MiR-206 (non-critical: n = 45, M = 0.2, IQR = 0.01–30.8; critical COVID-19: n = 55, M = 2.5, IQR = 0.1–59.8; * P = 0.03). MiR-205-5p (non-critical: n = 45, M = 0.9, IQR = 0.3–3.2; critical COVID-19: n = 55, M = 2.4, IQR = 0.6–6.1; *P = 0.03). MiR-22-3p (non-critical: n = 45, M = 0.9, IQR = 0.5–1.4; critical COVID-19: n = 55, M = 0.8, IQR = 0.6–2.1; P = 0.57). MiR-184 (non-critical: n = 45, M = 0.4, IQR = 0.04–83.2; critical COVID-19: n = 55, M = 0.3, IQR = 0.03–8.2; P = 0.69). MiR-4516 (non-critical: n = 45, M = 1.5, IQR = 0.1–4.6; critical COVID-19: n = 55, M = 1.0, IQR = 0.1–4.6; P = 0.83).
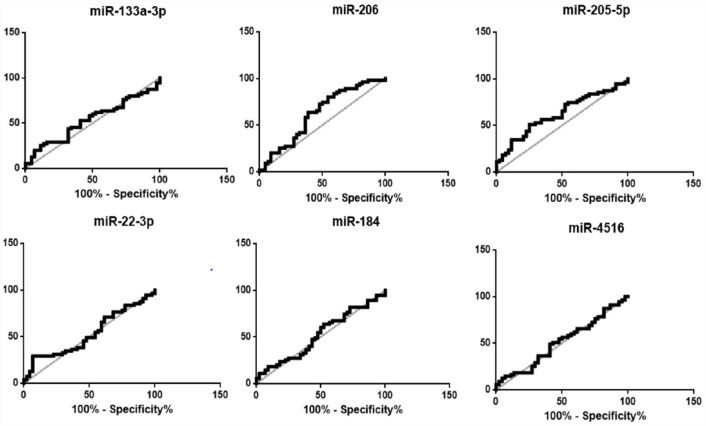
ROC curve analysis (Figure 4) showed that miR-205-5p was capable of discriminating patients at higher risk to develop a critical COVID-19, from non-critical disease (area under the curve [AUC] = 0.62, 95% confidence interval [CI] = 0.5–0.7, P = 0.03) with 73.3% of specificity and 50.0% of sensitivity. Moreover, patients with miR-205-5p serum expression levels superior to 2.37 (cutoff value = 2.37) were 3 times more likely to evolve to critical COVID-19 (odds ratio [OR] = 2.75; 95% CI = 1.18–6.43) than patients with lower miR-205-5p levels. Also, miR-206 presented a significant capacity to discriminate patients at higher risk to develop a critical form of the disease (AUC = 0.62, 95% CI = 0.5–0.7, P = 0.03) with 44.4% specificity and 79.6% sensitivity. In addition, patients who showed miR-206 serum expression levels superior than 0.10 (cutoff value = 0.10) were 3 times more likely to evolve to COVID-19 critical forms (OR = 3.12; 95% CI = 1.3–10.6) as compared to patients with lower levels of miR-206.
Figure 4.
ROC curve analysis of miR relative expressions in the expanded cohort of COVID-19 patients, considering critical COVID-19 as outcome.
ROC: receiver operating characteristic; COVID-19: coronavirus disease 2019; AUC: area under the curve; CI: confidence interval.
MiR-133a-3p (AUC = 0.53, 95% CI = 0.4–0.6, P = 0.54); MiR-206 (AUC = 0.62, 95% CI = 0.5–0.7, P = 0.03); MiR-205-5p (AUC = 0.62, 95% CI = 0.5–0.7, P = 0.03); MiR-22-3p (AUC = 0.52, 95% CI = 0.4–0.6, P = 0.57); MiR-184 (AUC = 0.52, 95% CI = 0.4–0.6, P = 0.6); MiR-4516 (AUC = 0.51, 95% CI = 0.4–0.6, P = 0.83).
Association between differentially expressed miRs and fatal COVID-19
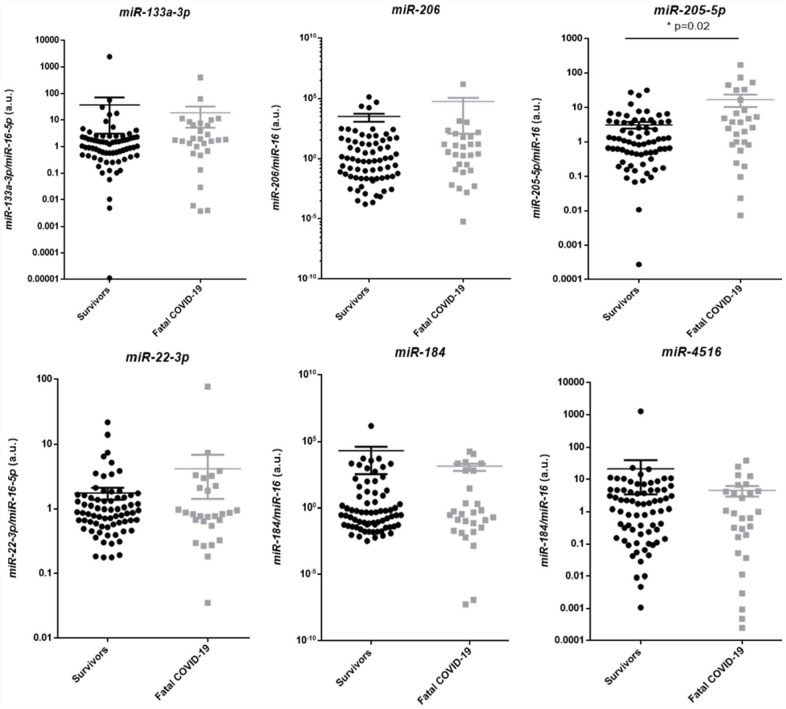
From the six dysregulated miRs detected in the screening cohort, only miR-205-5p remained differentially expressed in fatal COVID-19 compared to the survivors (non-critical and critical) when the expand cohort was tested. The expression of miR-205-5p was higher in fatal COVID-19 (n = 28, M = 3.2, IQR = 0.8–14.6) compared to survivors (n = 72, M = 1.0, IQR = 0.4–3.7, P = 0.03), while the relative expression of miR-133a-3p, miR-206, miR-22-3p, miR-184, and miR-4516 was similar between groups (Figure 5).
Figure 5.
miR validation in the expanded cohort of COVID-19 patients, comparing survivors with fatal COVID-19 patients.
miR: microRNA; COVID-19: coronavirus disease 2019; IQR: interquartile range; n: absolute number; M: median.
MiR-133a-3p (survivors: n = 72, M = 1.05, IQR = 0.5–2.3; fatal COVID-19: n = 28, M = 1.8, IQR = 0.6–7.2; P = 0.1). MiR-206 (survivors: n = 72, M = 0.7, IQR = 0.02–32.9; fatal COVID-19: n = 28, M = 3.1, IQR = 0.1–64.0; P = 0.24). MiR-205-5p (survivors: n = 72, M = 1.0, IQR = 0.4–3.7; fatal COVID-19: n = 28, M = 3.2, IQR = 0.8–14.6; *P = 0.02). MiR-22-3p (survivors: n = 72, M = 0.9, IQR = 0.6–1.5; fatal COVID-19: n = 28, M = 0.8, IQR = 0.6–2.2; P = 0.96). MiR-184 (survivors: n = 72, M = 0.30, IQR = 0.04–13.4; fatal COVID-19: n = 28, M = 0.33, IQR = 0.02–484.1, P = 0.94). MiR-4516 (survivors: n = 72, M = 1.7, IQR = 0.2–4.6; fatal COVID-19: n = 28, M = 0.7, IQR = 0.1–4.3; P = 0.43).
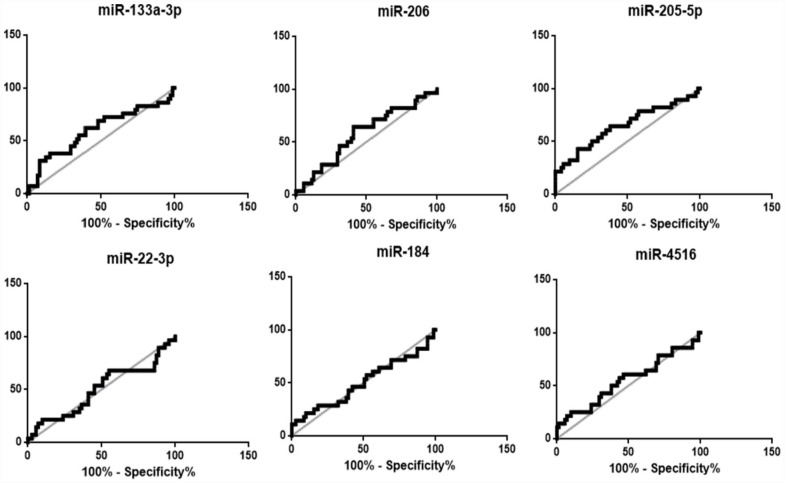
When death is considered as the outcome, ROC curve analysis showed that miR-205-5p levels upon hospital admission were capable of discriminating patients who died due to COVID-19 complications from survivors (AUC = 0.70, IQR = 0.6–0.8, P = 0.002), with 84.5% specificity and 42.9% sensitivity (Figure 6). Patients with miR-205-5p serum expression levels superior to 4.6 (cutoff value = 4.4) had around 4 times increased risk to die due to COVID-19 complications as compared to those with lower miR-205-5p levels (OR = 4.09, 95% CI = 1.52–10.97).
Figure 6.
ROC curve analysis of miRNA relative expressions in the expanded cohort of COVID-19 patients, considering fatal COVID-19 as outcome.
ROC: receiver operating characteristic; COVID-19: coronavirus disease 2019; AUC: area under the curve; CI: confidence interval.
MiR-133a-3p (AUC = 0.61, 95% CI = 0.5–0.7, P = 0.08); MiR-206 (AUC = 0.57, 95% CI = 0.4–0.7, P = 0.25); MiR-205-5p (AUC = 0.70, 95% CI = 0.6–0.8, P = 0.002**); MiR-22-3p (AUC = 0.52, 95% CI = 0.4–0.6, P = 0.75); MiR-184 (AUC = 0.54, 95% CI = 0.4–0.7, P = 0.5); MiR-4516 (AUC = 0.53, 95% CI = 0.4–0.6, P = 0.63).
Association between demographic and clinical characteristics of COVID-19 patients and the relative expression of miR-205-5p and miR-206
To investigate whether circulating miR-205-5p and miR-206 were independently associated with an increased risk of critical COVID-19 or death, we analyzed the correlation between patient age, sex, previous diseases, and miR-205-5p or miR-206 expression levels. No significant correlation was found between age and levels of miR-205-5p (correlation coefficient [R] = 0.1, P = 0.3) or miR-206 (R = 0.02, P = 0.8). Also, miR-205-5p levels were similar between male (n = 57, M = 0.9, IQR = 0.3–4.2) and female patients (n = 43, M = 1.4, IQR = 0.6–5.0; P = 0.23), and between patients without comorbidities (n = 30, M = 1.3, IQR = 0.5–3.6) and those with previous diseases (n = 70, M = 1.1, IQR: 0.4–5.5; P = 0.64). Similar results were found for miR-206 (male: n = 57, M = 1.5, IQR = 0.02–25.6; female: n = 43, M = 0.7, IQR = 0.05–52.3; P = 0.68 / no previous comorbidities: n = 30, M = 5.3, IQR = 0.1–67.1; previous comorbidities: n = 70, M = 0.9, IQR: 0.02–21.2; P = 0.39), which may indicate that basal conditions did not affect these miR serum expression levels.
In silico prediction of miR-205-5p target genes
As miR-205-5p expression levels were associated with critical and fatal COVID-19, an in silico analysis was performed to investigate possible biological pathways related to investigate the possible biological role of this miRNA on patients’ aggressive outcomes. We found that miR-205-5p may regulate over 400 different genes, most of them involved in molecular pathways of inflammation, immunological response, viral diseases, cellular metabolism, and cardiovascular risk factors. The results of gene enrichment analysis are shown in Figure 7.
Figure 7.
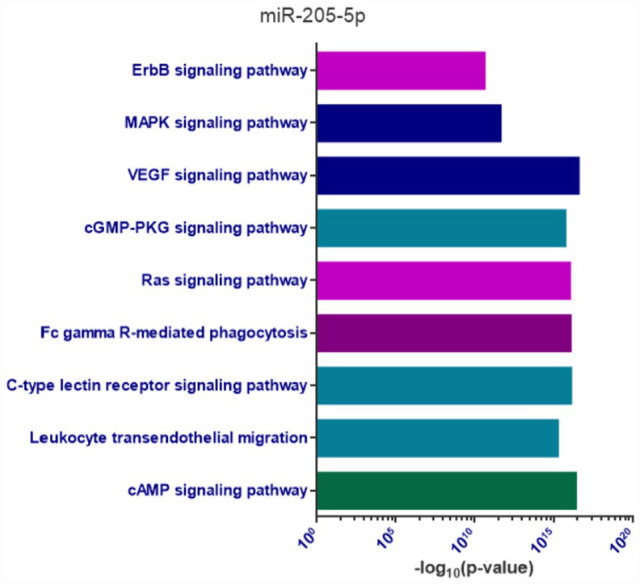
Gene enrichment analysis of miR-205-5p predict targets.
In this figure, biological pathways enriched (P < 0.05) by the predicted targets of miR-205-5p are represented, emphasizing processes related to inflammation, immunological response, other viral diseases, cardiovascular risk factors, and cellular metabolism.
Discussion
In this study, we evaluated the association between serum levels of miRNAs and COVID-19 severity in hospitalized patients. Compared to non-critical survivors, patients who needed intensive support to recover from COVID-19-related complications presented higher expression of circulating miR-205-5p and miR-206 upon hospital admission. The increased expression of miR-205-5p was also linked to a high mortality risk. Moreover, patients’ baseline characteristics apparently did not affect the expression of these miRs, which may indicate that circulating miR-205-5p and miR-206 expression levels could be a potential independent predictor of COVID-19 outcome in the beginning of the hospitalization period.
In the last 10 years, levels of miRs in peripheral blood samples have been pointed as regulators of gene expression and intercellular messengers in pathological states. In previous studies, miR-205-5p was described as a regulator of the angiogenic process in non-small cell and squamous lung cancer progression, and also as an influencer of the inflammatory responses in airway diseases.16–19
A decade ago, Suojalehto et al. 17 demonstrated that patients with symptomatic allergic rhinitis and asthma presented higher expression of miR-205-5p in nasal mucosa compared to non-symptomatic patients. These high levels of miR-205-5p were significantly correlated with an increased release of the cytokines interleukin (IL)-4, IL-5, and IL-13 by the activation of T-helper 2 (Th2) immune response, which may indicate that miR-205-5p participates in the establishment of tissue inflammation and clinical deterioration. In line with these findings, a recent study showed that miR-205-5p knockdown provides symptomatic relief in a mice model of allergic rhinitis and also minimizes the production of pro-inflammatory cytokines in nasal mucosa. This decreased inflammatory response allied with the induced lower expression of miR-205-5p was attributed to the regulation of the B-cell lymphoma 6 (BCL6) by miR-205-5p activity. 20
BCL6 is a transcriptional regulator that acts mainly targeting the promoter regions of genes related to the immunological responses in T cells, B cells, macrophages, mast cells, and airway epithelial cells.20,21 As a consequence of BCL6 regulation, a suppressed Th2 response with less production of IL-4, IL-5, and IL-13 cytokines and IgE is observed, as well as a decrease in the amount of macrophages producing IL-1β and IL-18. 20 This negative regulation of the inflammation responses by BCL6 occurs mainly due to the inhibition of the inflammasome linked to the nucleotide-binding oligomerization domain–like receptor family pyrin domain-containing 3 (NLRP3). 20 In fact, the authors also observed that together with symptomatic relief and less expression of immunological markers, miR-205-5p knockdown results in higher BCL6 protein and mRNA levels while decreasing NLRP3 levels. 20 NLRP3 inflammasome is a complex of proteins that, once activated, induces the apoptosis-associated speck-like protein containing a CARD (ASC), an inflammasome adapter presented mainly in T cells and macrophage nucleus. As a consequence of ASC induction, the caspase-1 cleavage may occur, enhancing IL-1β and IL-18 maturation, which supports the Th2 immunological response. 22 Also, this inflammatory effect related to the NLRP3 activation was already described in bronchus and epithelial mucous cells of asthmatic patients as an important factor related to lung inflammation and damage. 23
In the context of COVID-19, significant evidence supports the NLRP3 inflammasome activation as an essential step in developing tissue inflammatory response to SARS-CoV-2 infection in the lungs.24–27 The main hypothesis is that the N viral protein can directly bind to the NLRP3 complex, starting the release of IL-1β that culminates in the cytokine storm. In fact, both levels of NLRP3 proteins and IL-1β were found significantly increased in patients who presented SARS or multiple organ failure due to the infection. 25 Mice models of COVID-19-related NLPR3 activation showed that this process could be crucial during inflammation development and closely related to pulmonary injury, hypercoagulability state, and apoptotic signalization of the infected cells.24–26 The same events were also observed in other coronaviruses. 28
Other studies have shown that genetic variants of the NLPR3 inflammasome can influence the individual response to SARS-CoV-2 infection, which emphasizes that the activation of NLPR3 plays a key role during the inflammatory response.29,30 Yet, this is the first time that the highly expressed circulating miRNA was proposed as a positive regulator of NLPR3 during COVID-19 due to the inhibition of the BCL6 repressor, and as a biomarker of COVID-19 severity.
In addition to the pro-inflammatory effect of miR-205-5p by increasing NLPR3 expression, this miR has been previously implicated in antiangiogenic effects in diabetes-induced limb ulcer. 30 High levels of circulating miR-205-5p have been found to decrease protein levels of vascular endothelial growth factor A (VEGFA) through direct binding on its 3′UTR mRNA in “in vitro” and “in vivo” models. 31 VEGFA is one of the most important members of the VEGF family, 32 and its angiogenic activity is strongly induced by hypoxia. 33
However, in the context of critical COVID-19, the antiangiogenic role of miR-205-5p through the downregulation of VEGFA seems to be controversial. Despite being a key factor for tissue repair during inflammatory disease, the angiogenic process is considered an inductor of pulmonary endothelial damage in severe COVID-19. 34 Previous studies have demonstrated that patients who had severe lung injuries and organ failure presented high levels of VEGF family members in tissues (mainly VEGFA) and inflammatory cytokines. This induction of VEGFA tissue expression was attributed mainly to the cytokine storm process and to the COVID-19-related hypoxia state, which can induce a pro-angiogenic response. However, this angiogenic stimulus appears to be responsible for an increase of endothelial lung cell permeability that further impairs endothelium function and triggers tissue factor (TF) release, which leads to the establishment of a pro-thrombotic environment,35–38 once this interaction, in theory, could minimize the angiogenesis process and avoid organ damage. Thus, considering that our results showed that miR-205-5p is present in the serum of patients who presented non-fatal and fatal COVID-19, but significantly upregulated in the last cohort, we hypothesize that miR-205-5p could be involved in the COVID-19 pathophysiology both as an inducer of inflammation and as a cell regulator released in response to VEGFA over-expression. However, we cannot exclude the possibility that the increase in miR-205-5p expression may be a consequence of an increase in VEGFA levels.
In addition, we found that miR-206 was upregulated in serum samples of patients who needed intensive care to recover, but not in fatal COVID-19 or non-critical patients. miR-206 is consider a “miomiR” due to its enrichment in myocardial and vascular tissue. Also, several studies in cancer have demonstrated that miR-206 acts as an important antiangiogenic factor due to the downregulation of the VEGFA-linked axis.39–42 Although the antiangiogenic role of miR-206 in the COVID-19 context was not yet explored, we hypothesize that the increase in serum levels of this miR in patients who present critical, but non-fatal, forms of COVID-19 occurred as a compensatory mechanism, to control the exacerbated angiogenesis process provoked by the hyperinflammatory state caused by SARS-CoV-2 infection. Yet, further investigation is needed to clarify this point.
Our study has some limitations that should be highlighted. Although our results showed an association between high levels of circulating miR-205-5p and miR-206 upon hospital admission and increased risk of adverse outcomes, the potential of these miRs as predictive clinical biomarkers has to be confirmed in other cohorts. Besides, the post-transcriptional regulation of molecular pathways involved in COVID-19 pathophysiology by miR-205-5p activity remains to be proved by functional assays approaches, and once all our data were related to circulating miRNAs, the expression of miR-205-5p at tissue levels during the disease remains unknown. Finally, all demographic and clinical data reported in this article were obtained from medical records.
In conclusion, our study demonstrated that circulating miR-205-5p and miR-206 expression levels are capable of discriminating patients at higher risk to develop critical or fatal COVID-19 upon hospital admission. These results also indicate that epigenetic mechanisms may regulate the immunological response in COVID-19.
Footnotes
Authors’ contributions: All authors participated in the design, interpretation of the studies, and analysis of the data and review of the manuscript; CdOV conducted the experiments.
The author(s) declared no potential conflicts of interest with respect to the research, authorship, and/or publication of this article.
Ethical approval: This study was approved by the Local Research Ethics Committee (CAAE 30648520.6.0000.5404).
Funding: The author(s) disclosed receipt of the following financial support for the research, authorship, and/or publication of this article: This work was supported by the São Paulo Research Foundation – FAPESP (grant numbers: 2020/07922-4; 2020/12630-2), FAEPEX – University of Campinas (grant number: 519292/ 232120), and National Council for Scientific and Technological Development (CNPq) (grant numbers: 406990/2021-2; 308452/2021-6).
ORCID iDs: Camila de Oliveira Vaz  https://orcid.org/0000-0002-4860-1054
https://orcid.org/0000-0002-4860-1054
Fernanda Andrade Orsi  https://orcid.org/0000-0002-7908-9073
https://orcid.org/0000-0002-7908-9073
References
- 1. Lu H, Stratton CW, Tang YW. Outbreak of pneumonia of unknown etiology in Wuhan, China: the mystery and the miracle. J Med Virol 2020;92:401–2 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Zhu N, Zhang D, Wang W, Li X, Yang B, Song J, Zhao X, Huang B, Shi W, Lu R, Niu P, Zhan F, Ma X, Wang D, Xu W, Wu G, Gao GF, Tan W, China Novel Coronavirus Investigating and Research Team. A novel coronavirus from patients with pneumonia in China. 2019, N Engl J Med 2020;382:727–33 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Fernandes Q, Inchakalody VP, Merhi M, Mestiri S, Taib N, Moustafa Abo El-Ella D, Bedhiafi T, Raza A, Al-Zaidan L, Mohsen MO, Yousuf Al-Nesf MA, Hssain AA, Yassine HM, Bachmann MF, Uddin S. Emerging COVID-19 variants and their impact on SARS-CoV-2 diagnosis, therapeutics and vaccines. Ann Med 2022;54:524–40 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Hu B, Guo H, Zhou P, Shi Z-L. Characteristics of SARS-CoV-2 and COVID-19. Nat Rev Microbiol 2021;19:141–54 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Zhou F, Yu T, Du R, Fan G, Liu Y, Liu Z, Xiang J, Wang Y, Song B, Gu X, Guan L, Wei Y, Li H, Wu X, Xu J, Tu S, Zhang Y, Chen H, Cao B. Clinical course and risk factors for mortality of adult inpatients with COVID-19 in Wuhan, China: a retrospective cohort study. Lancet 2020;395:1054–62 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Bivona G, Agnello L. Biomarkers for prognosis and treatment response in COVID-19 patients 2021;41:540–8 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Lagos-Quintana M, Rauhut R, Lendeckel W, Tuschl T. Identification of novel genes coding for small expressed RNAs. Science 2001;294:853–8 [DOI] [PubMed] [Google Scholar]
- 8. Selbach M, Schwanhäusser B, Thierfelder N, Fang Z, Khanin R, Rajewsky N. Widespread changes in protein synthesis induced by microRNAs. Nature 2008;455:58–63 [DOI] [PubMed] [Google Scholar]
- 9. Pinilla L, Benitez ID, González J, Torres G, Barbé F, de Gonzalo-Calvo D. Peripheral blood microRNAs and the COVID-19 patient: methodological considerations, technical challenges and practice points 2021;18:688–95 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Biswas S, Haleyurgirisetty M, Lee S, Hewlett I, Devadas K. Development and validation of plasma miRNA biomarker signature panel for the detection of early HIV-1 infection. EBioMedicine 2019;43:307–16 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Weis A, Marquart L, Calvopina DA, Genz B, Ramm GA, Skoien R. Serum MicroRNAs as biomarkers in hepatitis C: preliminary evidence of a MicroRNA panel for the diagnosis of hepatocellular carcinoma. Int J Mole Sci 2019;20:864. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Amini-Farsani Z, Yadollahi-Farsani M, Arab S, Forouzanfar F, Yadollahi M, Asgharzade S. Prediction and analysis of microRNAs involved in COVID-19 inflammatory processes associated with the NF-kB and JAK/STAT signaling pathways. Int Immunopharmacol 2021;100:108071. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Marchese V, Crosato V, Gulletta M, Castelnuovo F, Cristini G, Matteelli A, Castelli F. Strongyloides infection manifested during immunosuppressive therapy for SARS-CoV-2 pneumonia. Infection 2021;49:539–42 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Mallick B, Ghosh Z, Chakrabarti J. MicroRNome analysis unravels the molecular basis of SARS infection in bronchoalveolar stem cells. PLoS ONE 2009;4:e7837 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Pfaffl MW. A new mathematical model for relative quantification in real-time RT-PCR. Nucleic Acids Res 2001;29:e45 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Kuang Y, Hu B, Huang M, Zhao S, Wu X, Zhang M, Xie Z. Phosphatidylethanolamine-binding protein 1 (PEBP1) mediates the regulatory role of microRNAs (miRNAs)-205-5p in degranulation and histamine release. Bioengineered 2022;13:13341–51 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Suojalehto H, Toskala E, Kilpeläinen M, Majuri ML, Mitts C, Lindström I, Puustinen A, Plosila T, Sipilä J, Wolff H, Alenius H. MicroRNA profiles in nasal mucosa of patients with allergic and nonallergic rhinitis and asthma. Int Forum Allergy Rhinol 2013;3:612–20 [DOI] [PubMed] [Google Scholar]
- 18. Fan X, Zou X, Liu C, Liu J, Peng S, Zhang S, Zhou X, Wang T, Geng X, Song G, Zhu W. Construction of the miRNA-mRNA regulatory networks and explore their role in the development of lung squamous cell carcinoma. Front Mol Biosci 2022;9:888020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Fan L, Li B, Li Z, Sun L. Identification of autophagy related circRNA-miRNA-mRNA-subtypes network with radiotherapy responses and tumor immune microenvironment in non-small cell lung cancer. Front Genet 2021;12:730003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Zhang S, Lin S, Tang Q, Yan Z. Knockdown of miR‑205‑5p alleviates the inflammatory response in allergic rhinitis by targeting B‑cell lymphoma 6. Mol Med Rep 2021;24:818. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Koh B, Ulrich BJ, Nelson AS, Panangipalli G, Kharwadkar R, Wu W, Xie MM, Fu Y, Turner MJ, Paczesny S, Janga SC, Dent AL, Kaplan MH. Bcl6 and Blimp1 reciprocally regulate ST2(+) Treg-cell development in the context of allergic airway inflammation. J Allergy Clin Immunol 2020;146:1121–369 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Kelley N, Jeltema D, Duan Y, He Y. The NLRP3 inflammasome: an overview of mechanisms of activation and regulation. Int J Mol Sci 2019;20:3328. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Birrell MA, Eltom S. The role of the NLRP3 inflammasome in the pathogenesis of airway disease. Pharmacol Ther 2011;130:364–70 [DOI] [PubMed] [Google Scholar]
- 24. Freeman TL, Swartz TH. Targeting the NLRP3 inflammasome in severe COVID-19. Front Immunol 2020;11:1518. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Pan P, Shen M, Yu Z, Ge W, Chen K, Tian M, Xiao F, Wang Z, Wang J, Jia Y, Wang W, Wan P, Zhang J, Chen W, Lei Z, Chen X, Luo Z, Zhang Q, Xu M, Li G, Li Y, Wu J. SARS-CoV-2 N protein promotes NLRP3 inflammasome activation to induce hyperinflammation. Nat Commun 2021;12:4664. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Potere N, Del Buono MG, Caricchio R, Cremer PC, Vecchié A, Porreca E, Dalla Gasperina D, Dentali F, Abbate A, Bonaventura A. Interleukin-1 and the NLRP3 inflammasome in COVID-19: pathogenetic and therapeutic implications. EBioMedicine 2022;85:104299. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Rodrigues TS, De Sá KSG, Ishimoto AY, Becerra A, Oliveira S, Almeida L, Gonçalves AV, Perucello DB, Andrade WA, Castro R, Veras FP, Toller-Kawahisa JE, Nascimento DC, De Lima MHF, Silva CMS, Caetite DB, Martins RB, Castro IA, Pontelli MC, De Barros FC, Do Amaral NB, Giannini MC, Bonjorno LP, Lopes MIF, Santana RC, Vilar FC, Auxiliadora-Martins M, Luppino-Assad R, De Almeida SCL, De Oliveira FR, Batah SS, Siyuan L, Benatti MN, Cunha TM, Alves-Filho JC, Cunha FQ, Cunha LD, Frantz FG, Kohlsdorf T, Fabro AT, Arruda E, de Oliveira RDR, Louzada-Junior P, Zamboni DS. Inflammasomes are activated in response to SARS-CoV-2 infection and are associated with COVID-19 severity in patients. J Exp Med 2021;218:e20201707 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Vora SM, Lieberman J. Inflammasome activation at the crux of severe COVID-19 2021;21:694–703 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Maes M, Tedesco Junior WLD, Lozovoy MAB, Mori MTE, Danelli T. In COVID-19, NLRP3 inflammasome genetic variants are associated with critical disease and these effects are partly mediated by the sickness symptom complex: a nomothetic network approach 2022;27:1945–55 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. De Sá NBR, Neira-Goulart M. Inflammasome genetic variants are associated with protection to clinical severity of COVID-19 among patients from Rio de Janeiro, Brazil 2022;2022:9082455. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Liu J, Wang J, Fu W, Wang X, Chen H, Wu X, Lao G, Wu Y, Hu M, Yang C, Yan L, Ren M. MiR-195-5p and miR-205-5p in extracellular vesicles isolated from diabetic foot ulcer wound fluid decrease angiogenesis by inhibiting VEGFA expression. Aging 2021;13:19805–21 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Ferrara N, Adamis AP. Ten years of anti-vascular endothelial growth factor therapy. Nat Rev Drug Discov 2016;15:385–403 [DOI] [PubMed] [Google Scholar]
- 33. Semenza GL. Angiogenesis in ischemic and neoplastic disorders. Annu Rev Med 2003;54:17–28 [DOI] [PubMed] [Google Scholar]
- 34. Xu S-W, Ilyas I, Weng J-P. Endothelial dysfunction in COVID-19: an overview of evidence, biomarkers, mechanisms and potential therapies. Acta Pharmacolo Sin 2023;44:695–709 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. De Moraes CRP, De Borba Junior IT, De Lima F, Barbosa MS, Huber SC, Palma AC, Nunes TA, Ulaf RG, Ribeiro LC, Bernandes AF, Bombassaro B, Dertkigil SS, Moretti ML, Bizzacchi JA, Mansour E, Velloso LA, Maranhão Costa FT, Orsi FA, De Paula EV. Circulating levels of Ang/Tie2 and VEGF-a pathway mediators are associated with clinical severity, Endothelial barrier disruption and coagulation activation in COVID-19. Blood 2021;138:2073 [Google Scholar]
- 36. Goyal A, Prasad R, Goel P, Pal A, Prasad S, Rani I. An integrated approach of the potential underlying molecular mechanistic paradigms of SARS-CoV-2-mediated coagulopathy. Indian J Clin Biochem 2021;36:387–403 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Mayi BS, Leibowitz JA, Woods AT, Ammon KA, Liu AE. The role of neuropilin-1 in COVID-19 2021;17:e1009153 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Pons S, Fodil S, Azoulay E, Zafrani L. The vascular endothelium: the cornerstone of organ dysfunction in severe SARS-CoV-2 infection. Critical Care 2020;24:353. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Sun X, Luo L, Li J. LncRNA MALAT1 facilitates BM-MSCs differentiation into endothelial cells via targeting miR-206/VEGFA axis. Cell Cycle 2020;19:3018–28 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. De Oliveira A, Castanhole-Nunes MMU, Biselli-Chicote PM, Pavarino ÉC, Da Silva R, Da Silva RF, Goloni-Bertollo EM. Differential expression of angiogenesis-related miRNAs and VEGFA in cirrhosis and hepatocellular carcinoma. Arch Med Sci 2020;16:1150–7 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. Zheng X, Ma YF, Zhang XR, Li Y, Zhao HH, Han SG. Circ_0056618 promoted cell proliferation, migration and angiogenesis through sponging with miR-206 and upregulating CXCR4 and VEGF-A in colorectal cancer. Eur Rev Med Pharmacol Sci 2020;24:4190–202 [DOI] [PubMed] [Google Scholar]
- 42. Wang S, Ren L, Shen G, Liu M, Luo J. The knockdown of MALAT1 inhibits the proliferation, invasion and migration of hemangioma endothelial cells by regulating MiR-206 / VEGFA axis. Mol Cell Probes 2020;51:101540. [DOI] [PubMed] [Google Scholar]