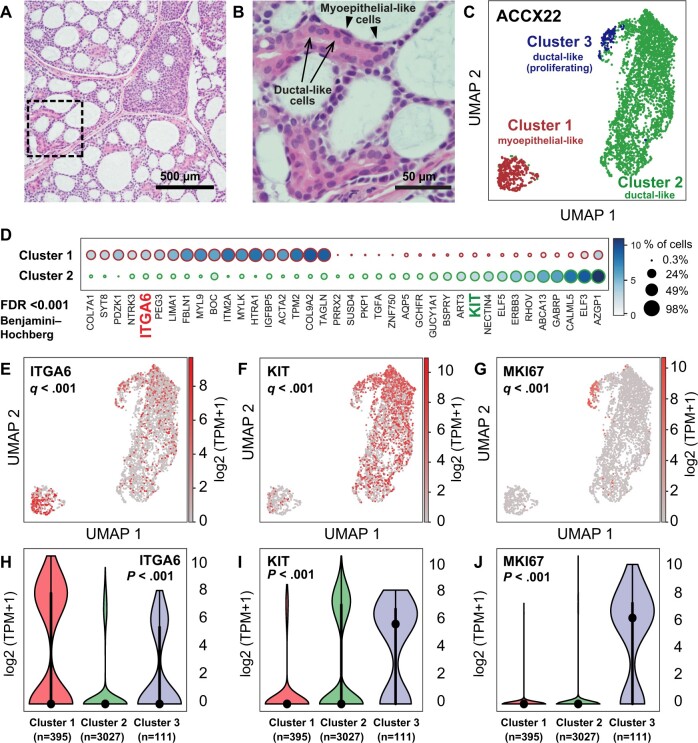
Figure 1.
Identification of surface markers for the differential purification of myoepithelial-like and ductal-like cell populations in human ACCs. A) Histological analysis of the ACCX22 human PDX line, confirming retention of a cribriform histology with pseudocyst formation, characteristic of well-differentiated (grade 1) ACCs. B) Magnification of the tissue area outlined in (A) (dashed box), demonstrating the presence of 1) ductal-like cells, characterized by abundant eosinophilic cytoplasm and arranged in ringlike structures (arrows); and 2) myoepithelial-like cells (arrowheads), characterized by spindle-shaped morphology and arranged to line pseudocysts. C) Visualization by Uniform Manifold Approximation and Projection (UMAP) of scRNA-seq data obtained from a purified preparation of human malignant cells (EpCAM+) sorted by FACS from the ACCX22 human PDX line. In the UMAP scatter plot, the 3 cell clusters identified as representing the most robust clustering solution (ie, as displaying the highest mean silhouette score following clustering based on the Leiden algorithm) were labeled with different colors and displayed clear visual separation. Based on differentially expressed genes (Supplementary Figure 3, available online), the 3 clusters were annotated as follows: cluster 1 = myoepithelial-like cells; cluster 2 = ductal-like cells; cluster 3 = proliferating ductal-like cells. D) List of genes identified as displaying a statistically significant difference in mean expression levels between cluster 1 (myoepithelial-like) and cluster 2 (ductal-like), based on a Student’s t test (2-tailed) adjusted for multiple comparisons (FDR < 0.001; Benjamini–Hochberg method). Among the differentially expressed genes are those encoding for 2 surface markers: ITGA6 (CD49f) and KIT (CD117). E-G) UMAP plots displaying gene-expression levels for ITGA6 (E), KIT (F), and the proliferation marker MKI67 (G); q values are based on a Student's t test (2-tailed), corrected for multiple comparisons (Benjamini–Hochberg method), as described in Supplementary Figure 3 (available online). H-J) Violin plots displaying the distribution of gene-expression levels for ITGA6 (H), KIT (I), and MKI67 (J) across the 3 cell clusters identified by scRNA-seq; P values are based on a Kruskal–Wallis H test. ACC = adenoid cystic carcinoma; FACS = fluorescence-activated cell sorting; FDR = false discovery rate; PDX = patient-derived xenograft; scRNA-seq = single-cell RNA sequencing; TPM = transcripts per million.