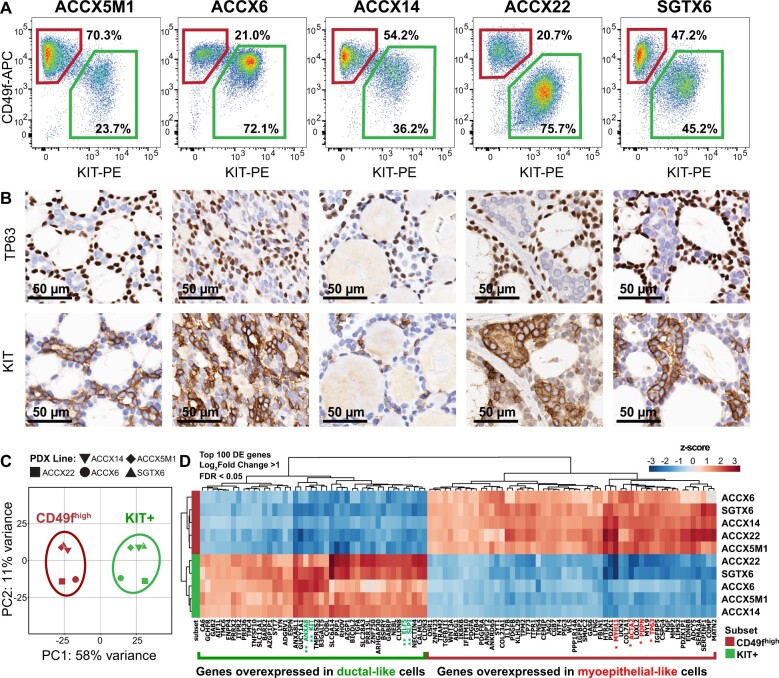
Figure 2.
Differential purification by flow cytometry of myoepithelial-like (CD49fhigh/KITneg) and ductal-like (CD49flow/KIT+) cells. A) Analysis by flow cytometry of CD49f and KIT surface expression in 5 human PDX lines representative of biphenotypic ACCs, enabling visual discrimination of 2 distinct populations of human malignant cells: CD49fhigh/KITneg (top-left gates) and CD49flow/KIT+ (bottom-right gates). B) Analysis by immunohistochemistry (IHC) of corresponding tumors, confirming mutually exclusive expression of TP63 (a myoepithelial marker) and KIT (a ductal marker). Scale bar: 50 µm. C) Principal component analysis (PCA) of RNA-seq data obtained from 5 autologous pairs of CD49fhigh/KITneg (CD49fhigh) and CD49flow/KIT+ (KIT+) cells, purified in parallel from 5 biphenotypic PDX lines of human ACCs (ACCX5M1, ACCX6, ACCX14, ACCX22, SGTX6). PCA was performed using the top 500 genes displaying the highest level of variance across the full 10-sample dataset. The 10 samples segregated into 2 distinct clusters, corresponding to their surface marker phenotype (CD49fhigh/KITneg vs CD49flow/KIT+) and separating along the first principal component (PC1). (D) Heatmap of the top 100 genes identified as differentially expressed (DE) between CD49fhigh/KITneg and CD49flow/KIT+ cells, after mean centering of gene-expression levels and hierarchical clustering of both genes and samples. Differentially expressed genes were defined as those with a more than twofold difference in mean expression levels between the 2 populations (log2 fold-change >1) that was considered to be statistically robust based on a Wald test corrected for multiple comparisons (FDR < 0.05; Benjamini–Hochberg method). Differentially expressed genes were ranked based on the P value from the Wald test. Asterisk labels indicate genes encoding for previously validated myoepithelial (*) and ductal (**) markers. ACC = adenoid cystic carcinoma; APC = allophycocyanin; DE = differentially expressed; FDR = false discovery rate; PC1 = first principal component; PC2 = second principal component; PDX = patient-derived xenograft; PE = phycoerythrin; RNA-seq = RNA sequencing.