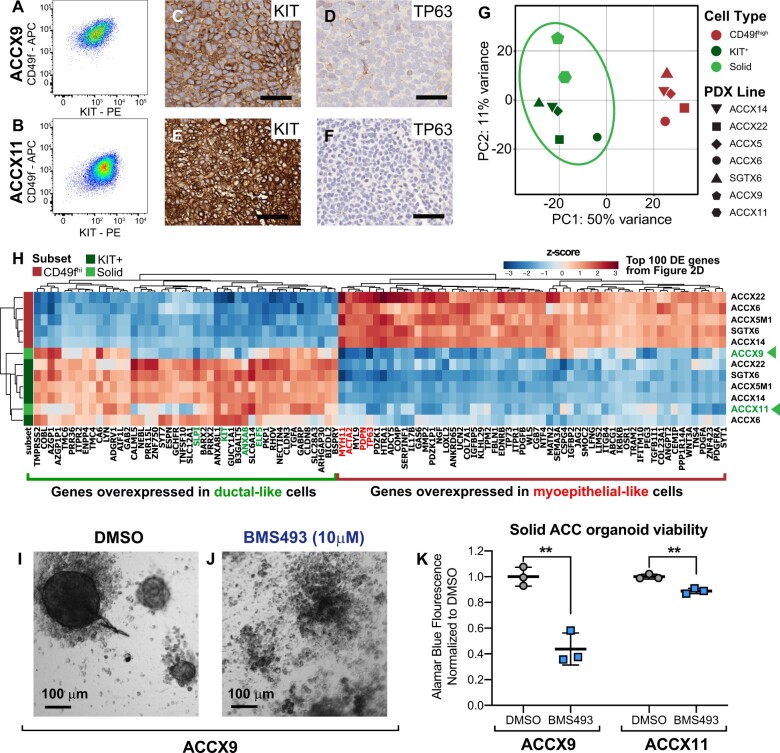
Figure 7.
Transcriptional profile and drug sensitivity of ACCs with solid histology. A-B) Analysis by flow cytometry of 2 PDX lines representative of human ACCs with solid histology ([A] ACCX9; [B] ACCX11) revealing a monophenotypic, ductal-like (CD49flow/KIT+) cell composition. C-F) Analysis by IHC of KIT and TP63 expression in ACCX9 and ACCX11 tumors, showing ubiquitous expression of the ductal-specific marker KIT (C, E) and complete loss of the myoepithelial-specific marker TP63 (D, F). Scale bars: 50 µm. G) Principal component analysis (PCA) of RNA-seq data from human ACCs, in which data from the 2 PDX lines with solid histology (ACCX9, ACCX11) are combined with those from the 5 autologous pairs of CD49fhigh/KITneg and CD49flow/KIT+ cells isolated from biphenotypic PDX lines (ACCX5M1, ACCX6, ACCX14, ACCX22, SGTX6). PCA was performed using the top 500 genes that displayed the highest level of variance across the full 12-sample dataset. H) Hierarchical clustering of RNA-seq data from human ACCs, based on the expression levels of the same list of 100 genes identified as differentially expressed (DE) between CD49fhigh/KITneg and CD49flow/KIT+ cells and reported in Figure 2, D. Solid ACCs clustered with CD49flow/KIT+ cells from biphenotypic tumors, irrespective of the method used to analyze their transcriptional profile. I-J) On visual inspection by conventional microscopy, ACCX9 organoids cultured for 1 week in the presence of BMS493 (10 µM) displayed widespread cell fragmentation, in contrast to organoids cultured with DMSO alone. K) Quantification of organoid viability using the alamarBlue assay, confirming the cytotoxic activity of BMS493 (10 µM) against PDX lines with solid histology (ACCX9, ACCX11). Error bars: mean +/- standard deviation; P values: Student’s t test (2-tailed; **P < .01). ACC = adenoid cystic carcinoma; APC = allophycocyanin; DMSO = dimethyl-sulfoxide; IHC = immunohistochemistry; PC1 = first principal component; PC2 = second principal component; PDX = patient-derived xenograft; PE = phycoerythrin; RNA-seq = RNA sequencing.