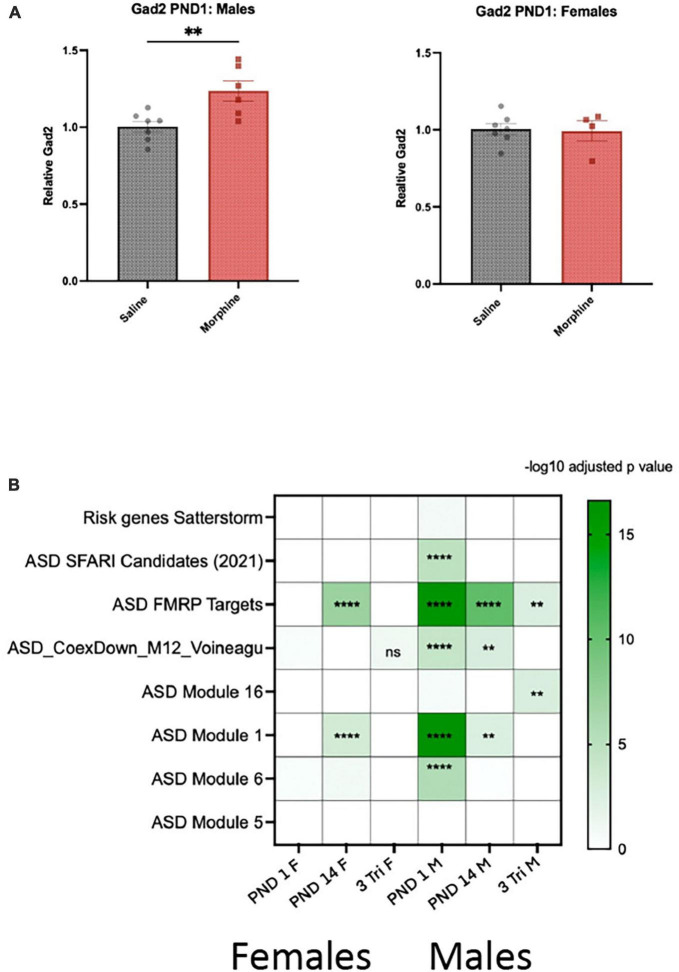
FIGURE 6.
Gene expression and enrichment in ASD datasets (A) Gad2 mRNA was assayed by qPCR in PND1 males and females exposed in utero to either morphine or saline (N = 7M, saline; 6M, morphine; 7Fm saline; 4F, morphine). 2-tail t-test, p < 0.01 for males, N.S. for females. (B) Heatmap showing the FDR for the overlap between DEGs identified in females and males and genes related to autism. Overlap was computed using Fisher’s exact test. Color indicates the -log10 FDR obtained after the correction with Benjamini-Hochberg method. Overlaid asterisks indicate the significance as following: **FDR < 0.01; ****FDR < 0.0001. “ASD Candidates SFARI (2021)”: ASD genetic risk factors from the SFARI database (gene scores 1 to 3 and syndromic); from Abrahams et al.; “Risk genes Satterstorm”: high confidence risk genes described by Satterstrom et al.;“ASD_FMRPTargets”: FMRP (Fragile X mental retardation protein) binding targets from Darnell et al.; “ASD Module 12”: ASD-associated co-expression module 12 and “ASD Module 16”: ASD-associated co-expression module 16 from Voineagu et al.; “ASD Module 1”: ASD-associated co-expression module 1 and “ASD Module 6”: ASD-associated co-expression module 6 and “ASD Module 5”: ASD-associated co-expression module 5 from Gupta et al.