Extended Data Figure 7. Synapse prediction sensitivity analysis.
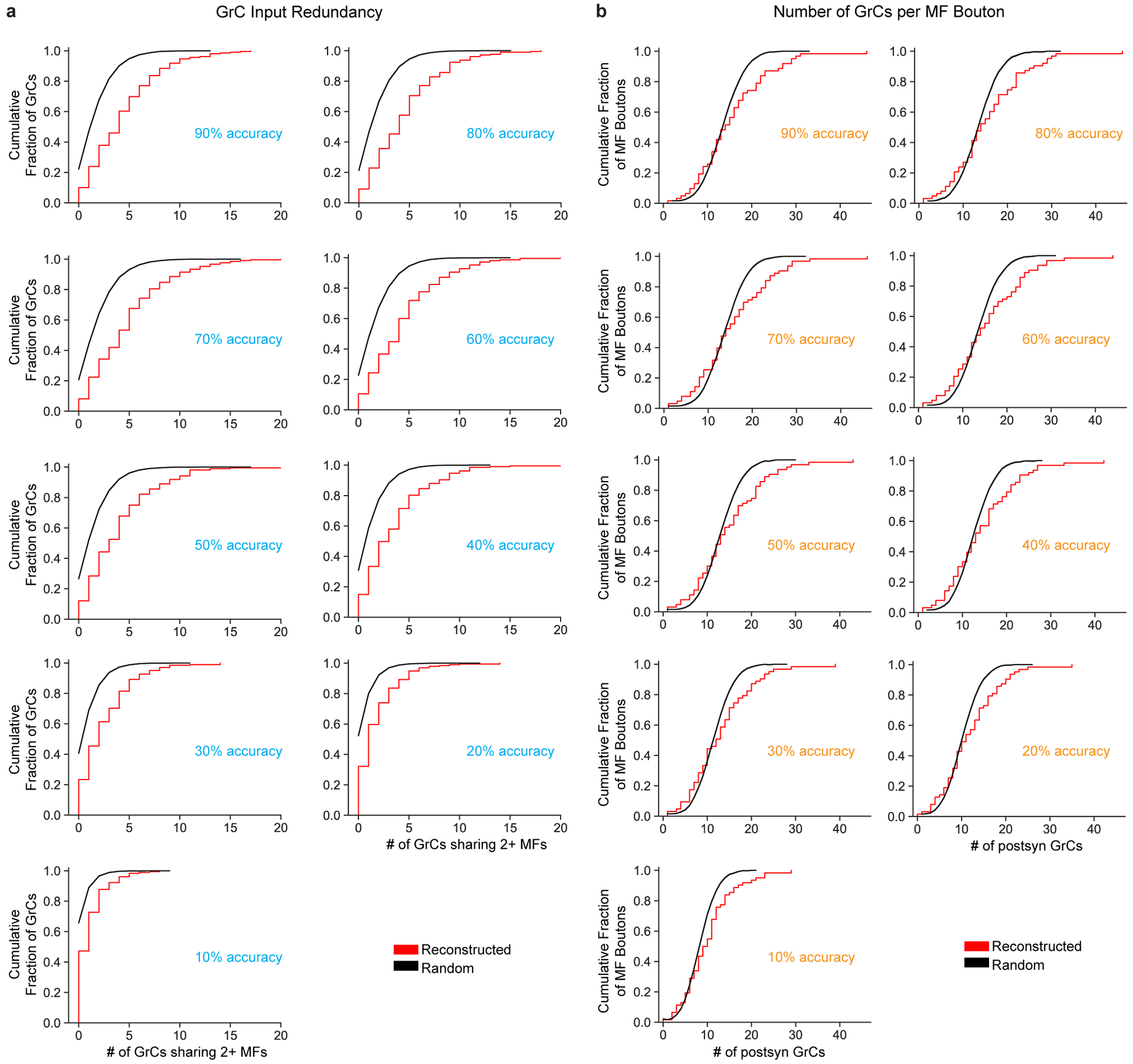
a, Cumulative distributions of MF bouton input redundancy as in Fig. 2c but across synapse prediction accuracies ranging from 90% to 10%. We artificially added false positives (FPs) and false negatives (FNs) to the network (Extended Data Fig. 2f) to achieve different accuracies (Methods). b, Cumulative distribution of postsynaptic GrCs per MF bouton as in Fig. 2e but across synapse prediction accuracies. As shown in a and b, we found that the results were consistent across models and only changed substantially when the FP/FN rates increased past 60%. We propose two reasons our results are robust across model prediction accuracies. First, MF-GrC connections are typically composed of multiple synapses (10 on average, Extended Data Fig. 4c). Since we used at least 3 synapses as a threshold for determining connectivity, even with significant missing, undetected synapses (eg. 50%), the remaining synapses are still reliable to reflect binary connectivity. Second, random and spurious false positive predictions are unlikely to coincide to cross the 3-synapse threshold. One interesting implication is that strongly selective features do not require perfect synapse prediction. This is consistent with connectomes in Drosophila where synapse prediction accuracy is ~60% and that connections are typically consist of multiple synapses which means that even with significant missing, undetected synapses (eg. 50%), the remaining synapses are sufficiently reliable to indicate connectivity.
