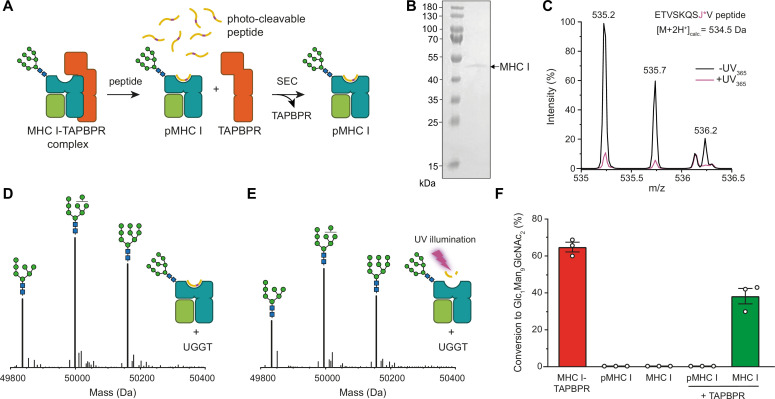
Figure 3. TAPBPR is indispensable for UGGT1-catalyzed reglucosylation of HLA-A*68:02.
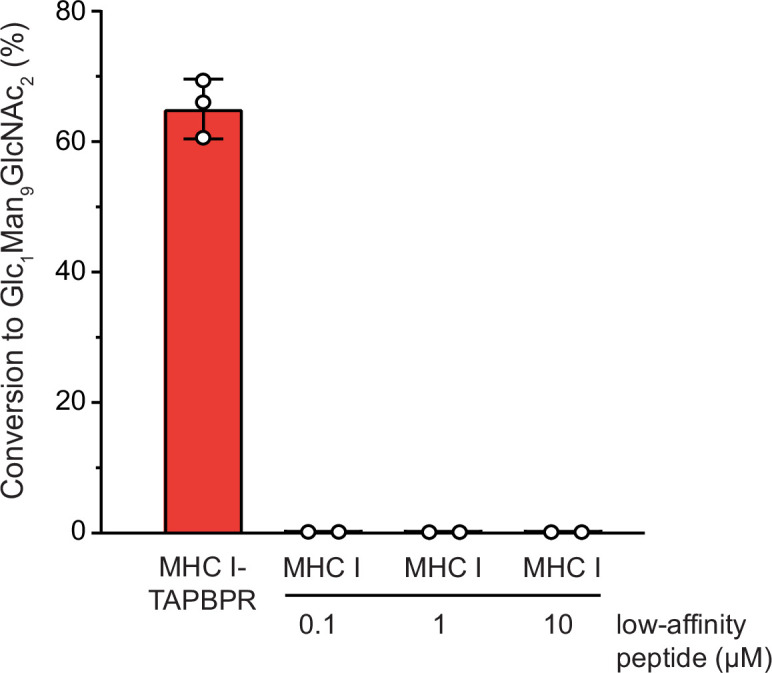
(A) Schematic representation of loading a photo-cleavable peptide onto Man9GlcNAc2-HLA-A*68:02 via pre-assembled MHC I-TAPBPR complexes. Peptide binding results in TAPBPR dissociation, permitting isolation of photo-conditional pMHC I complexes. (B) Non-reducing SDS-PAGE analysis of SEC-isolated pMHC I. (C) Isotope-resolved raw mass spectrum of the double-charged photo-cleavable peptide ETVSKQSJ*V (J*, 3-amino-3-(2-nitrophenyl)-propanoic acid) bound to HLA-A*68:02 before (black) and after (violet) UV illumination at 365 nm. (D) Reglucosylation experiment of peptide-loaded and (E) peptide-free HLA-A*68:02 (1 µM) in the absence of TAPBPR. Man9GlcNAc2-HLA-A*68:02 devoid of peptide was produced by photo-cleavage and release of peptide fragments prior to incubation with UGGT1 (600 nM). Shown are representative LC-MS analyses of data depicted in F. (F) Reglucosylation activity of UGGT1 (600 nM) towards TAPBPR-bound HLA-A*68:02 (1 µM), peptide-bound HLA-A*68:02 (pMHC I, 1 µM), and UV-illuminated HLA-A*68:02 lacking peptide (1 µM). If TAPBPR (6 µM) is added back to peptide-deficient MHC I (1 µM), recognition of MHC I as a substrate by UGGT1 is restored. The amount of produced Glc1Man9GlcNAc2 glycan was determined after 60 min for all reactions. Data represent mean ± SEM (n=3). Abbreviations: calc.: calculated; Da: dalton; kDa: kilodalton; SEC: size-exclusion chromatography.
Figure 3—figure supplement 1. SEC-MS analysis of photo-cleavable peptide bound to Man9GlcNAc2-HLA-A*68:02 before and after UV illumination.
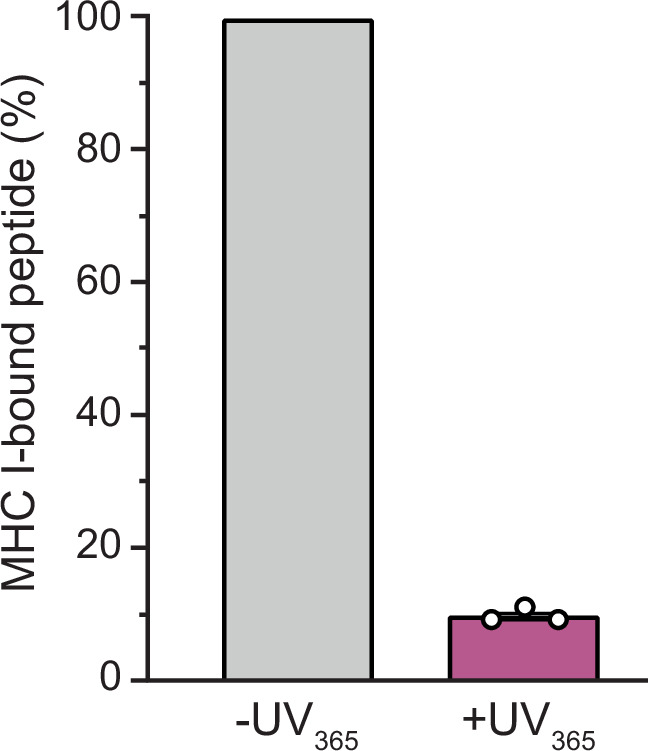
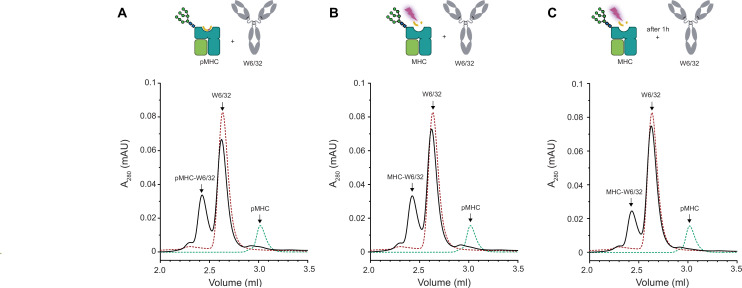
Figure 3—figure supplement 2. Conformation-specific antibody W6/32 binds to peptide-loaded and peptide-receptive β2m-HLA-A*68:02.
Figure 3—figure supplement 3. UGGT1-mediated reglucosylation is independent of the MHC I peptide-loading status.