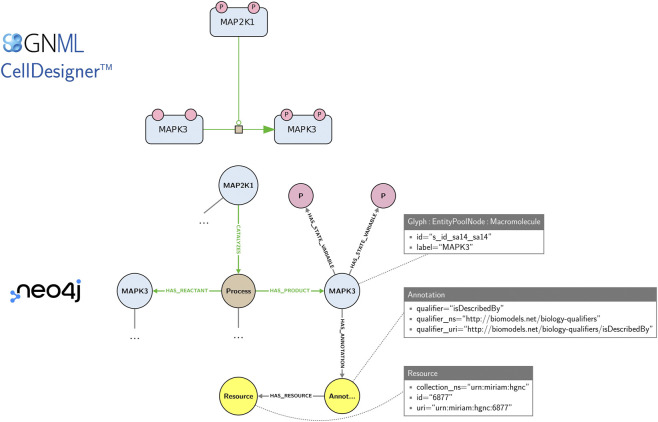
FIGURE 5.
An SBGN map and an excerpt of its corresponding Neo4j graph built using the StonPy software. SBGN nodes (e.g., proteins, states variables) are modelled using Neo4j nodes, while SBGN arcs (e.g., catalyses, production arcs) and relationships between concepts are modelled using Neo4j relationships (edges). Neo4j nodes are labelled (e.g., “Glyph”, “Macromolecule”) and may contain key-value pairs (e.g., the pair “label”/“MAPK3”). Additionally, annotations are stored in a structured form that can be easily queried. In the complete model (not shown), SBGN arcs are additionally modelled using Neo4j nodes, as they may contain SBGN nodes themselves.