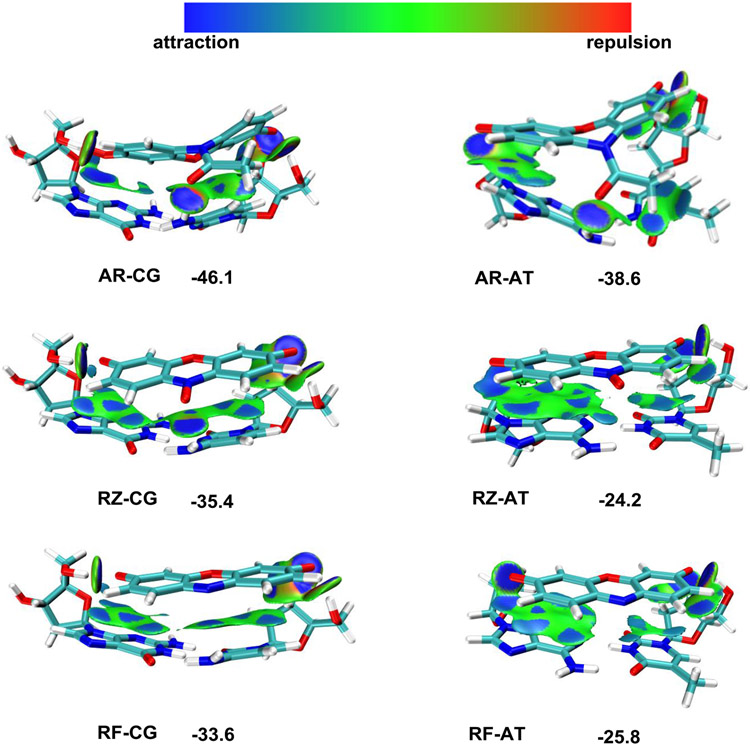
Figure 5.
DFT calculation of the binding between DNA base pairs and reaction substrates (AR, RZ, and RF). Optimized structure and IGMH (independent gradient model based on Hirshfeld partition) map (dginter = 0.05 a.u., isovalue = 0.004 a.u.) for AR-CG, AR-AT, RZ-CG, RZ-AT, RF-CG, and RF-AT. Values shown are binding energies in kcal/mol.