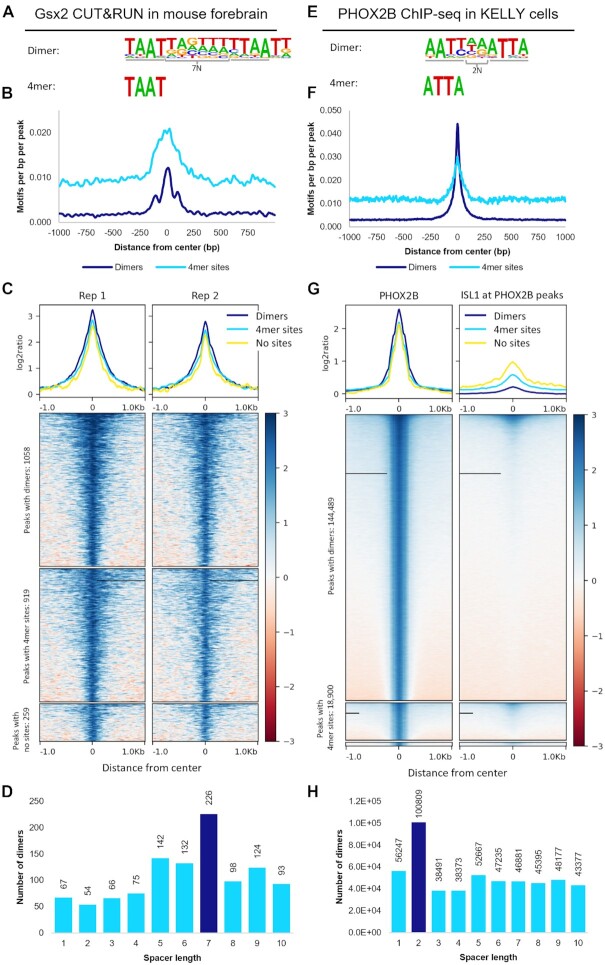
Figure 5.
Genomic binding assays reveal that predicted TFs bind cooperative dimer sites and non-cooperative monomer sites in vivo. (A) HT-SELEX revealed that GSX2 binds to a dimer site with TAAT 4mers spaced 7 bp apart. (B) Dimer motifs and the identified 4mers are highly prevalent in the centers of peaks called from mouse forebrain CUT&RUN assays. (C) Peaks containing Gsx2 dimer sites had a higher binding signal than peaks containing only 4mer sites or neither site, consistent with dimer sites either being bound more frequently or with higher stability than monomer sites. (D) Dimer sites consisting of 4mers 7bp apart are highly enriched compared to 4mers spaced by alternate distances. (E) PHOX2B HT-SELEX data enriched for a dimer motif consisting of 4mers 2bp apart and a monomer ATTA site. (F) These motifs are highly prevalent at the peak centers and (G) peaks containing a dimer site have increased binding compared to peaks with only a monomer site or lacking both types of sites. ISL1 does not bind many of the PHOX2B bound regions, which is consistent with these dimer sites being bound by PHOX2B homodimers rather than PHOX2B-ISL1 heterodimers. (H) 4mers spaced 2bp apart occurred nearly twice as frequently as the next most frequent spacing, consistent with the spacer specificity seen in cooperativity.