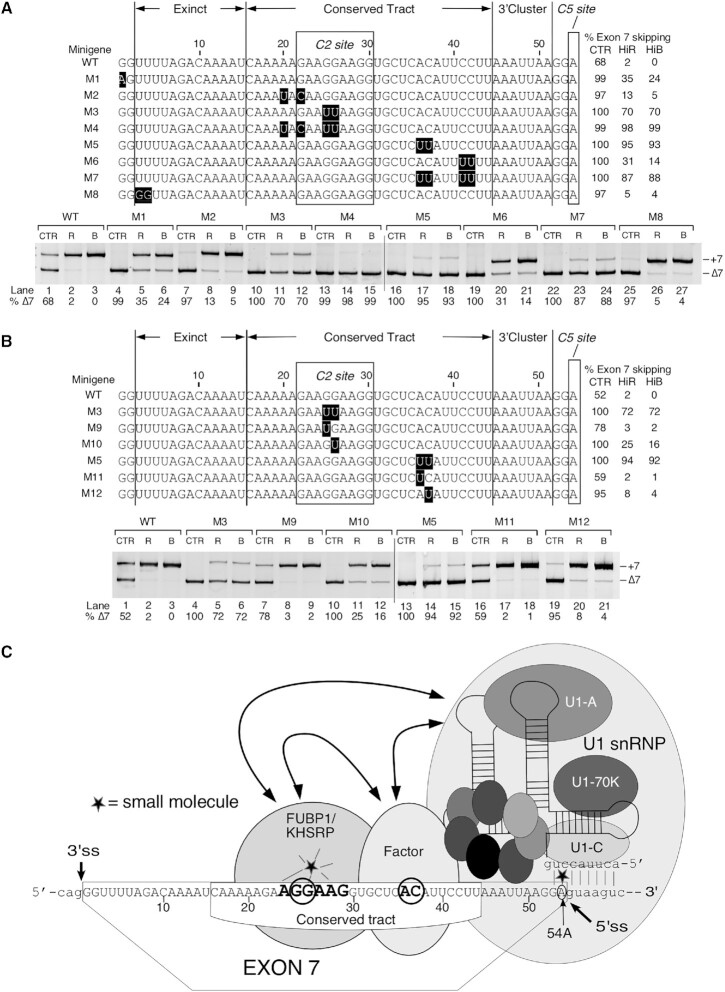
Figure 9.
Exonic mutations affect the ability of risdiplam and branaplam to promote exon 7 inclusion in transcripts generated from SMN2 minigenes. (A) Splicing of transcripts produced from SMN2ΔI6 minigenes carrying exonic mutations in HeLa cells. The sequence of the entire exon 7 for each minigene construct is shown in the upper panel. Minigene names are given on the left. Type of treatment and percentage of minigene exon 7 skipping are indicated on the right. Mutated nucleotides are highlighted in white on a black background. Three regions, Exinct, the Conserved tract and 3′ Cluster, shown to affect splicing of SMN exon 7 are indicated (13). C2 and C5 sites show where risdiplam C2 and C5 analogs interact with SMN exon 7 (45,46). A representative gel image showing the splicing pattern of SMN2 minigenes is given in the lower panel. Treatments and minigene names are indicated at the top of the gel image. Labeling is the same as in Figure 1B. (B) Splicing of SMN2 minigenes carrying single versus double mutations in HeLa cells. Labeling is the same as in (A). (C) Diagrammatic representation of the mechanism of small molecule-induced recruitment of U1 snRNP through GG and AC motifs. The sequence of SMN2 exon 7 and surrounding intronic sequences are given. Numbering is relative to the beginning of exon 7. Splicing factors are shown as shaded circles. Critical nucleotides for activity of risdiplam and branaplam are circled. FUBP1/KHSRP interacts with both the GG-rich motif and the small molecule. FUBP1/KHSRP may directly contact the downstream AC residues or help to recruit another factor that interacts with the downstream AC residues. Upstream RNA–protein and protein–protein interactions induced by the small molecule facilitate recruitment of U1 snRNP at the 5′ss that independently interacts with the small molecule and U1 snRNA.