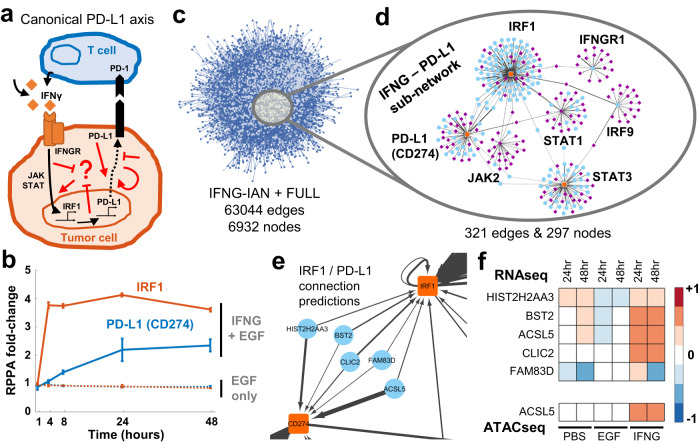
Fig. 5. Exploration of a single integrated association network reveals new links between IFNγ signaling and PD-L1.
a IFNγ secreted by T cells induces PD-L1 expression through JAK/STAT/IRF1 and other canonical pathways (black arrows). The PD-L1 on the cell surface then interacts with PD-1 on the immune cells to induce tumor cell death. However, the PD-1/PD-L1 therapy yields inter- and intratumor heterogeneous responses, and there is a need to identify new non-canonical targets (represented by red arrows). b The IFNγ induces IRF1 and PD-L1 production in MCF10A cells. Data shown are from three independent biological replicates and error bars represent the standard error of the mean. Source data are provided as a Source Data file. c The IFNG associations' network (IFNγ-IAN) is a data-driven large-scale network of connections. The associations are coalesced into gene-level nodes, and associations with greater than 0.01 absolute value are shown. d The sub-network of the IFNγ – PD-L1 relationship is significantly smaller than the IFNγ-IAN. The sub-network is generated by filtering for 14 genes and has seven hubs (PD-L1 (CD274), IFNGR1, IRF1, IRF9, JAK2, STAT1, STAT3) connected to 290 other genes. Node colors represent; blue:RNAseq, purple:ATACseq, and orange:RPPA and the edge widths correlate with the magnitude of the association coefficients. e A closer look at the connections between IRF1 and PD-L1 (CD274) genes shows a breadth of different functional genes. f Compared to no or EGF-only stimulation, the connecter genes are upregulated (RNAseq) in IFNγ stimulated condition. Additionally, the ACSL5 peak is more accessible similar to the canonical IFNγ downstream genes (ATACseq). The data shown are the average values from three independent biological replicates. Source data are provided as a Source Data file.