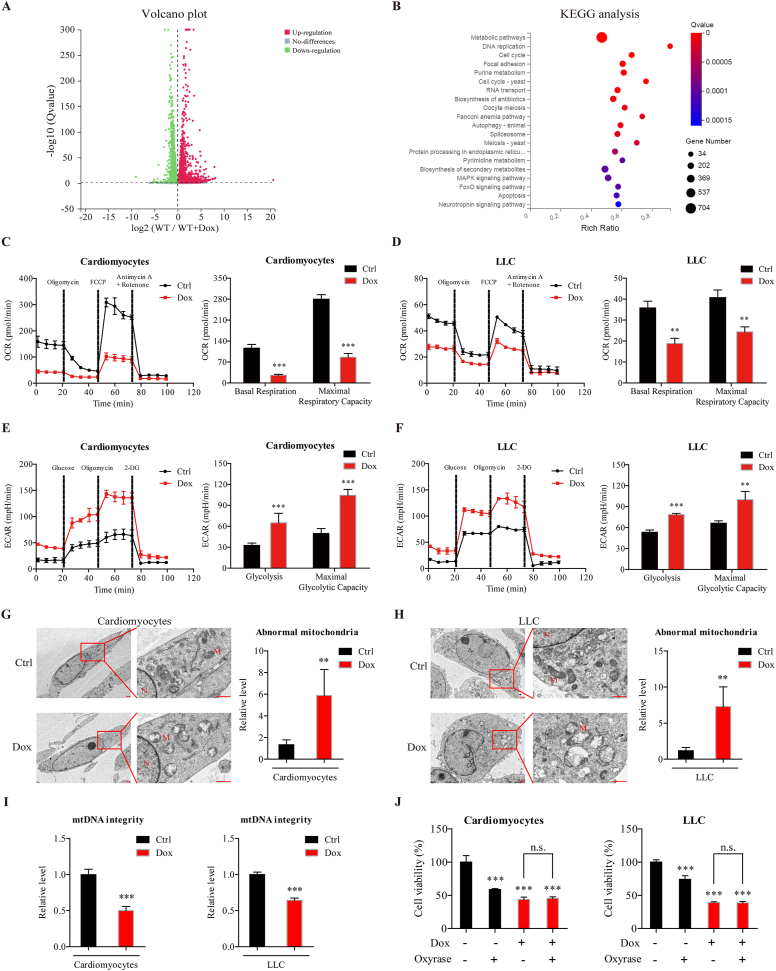
Figure 5.
Dox impairs mitochondrial respiration and mtDNA integrity to induce cytotoxicity in cardiomyocytes and cancer cells. (A) RNA sequencing was performed on primary cardiomyocytes isolated from WT mice, after treated with Dox (0.5 μmol/L) or solvent for 24 h. Differentially expressed genes are shown in volcano plots. (B) Kyoto Encyclopedia of Genes and Genomes (KEGG) pathway analysis of differentially expressed genes. (C, D) The oxygen consumption rate (OCR) was measured by a Seahorse XF96 extracellular-flux analyzer after cardiomyocytes and LLC cells were treated with Dox (0.5 μmol/L) for 12 h (n = 3 different experiments). (E, F) The extracellular acidification rate (ECAR) was measured by a Seahorse XF96 extracellular-flux analyzer after cardiomyocytes and LLC cells were treated with Dox (0.5 μmol/L) for 12 h (n = 3 different experiments). (G, H) Representative electron microscopic images of cardiomyocytes and LLC cells after Dox treatment (0.5 μmol/L) for 24 h are shown. Scale bar = 1 μm. The number of abnormal mitochondria was quantified blindly in 5 images from different fields. (I) Mitochondrial DNA (mtDNA) integrity was determined after cardiomyocytes and LLC cells were treated with Dox (0.5 μmol/L) for 24 h (n = 3 different experiments). (J) Cell viability was measured by the CCK-8 assay after cardiomyocytes and LLC cells were treated with oxyrase and/or Dox (0.5 μmol/L) for 24 h (n = 3 different experiments). The data are expressed as the mean ± standard deviation. Two-tailed unpaired t tests were performed to determine significant differences in C–I. One-way ANOVA followed by Bonferroni multiple comparisons were performed to determine significant differences in J. ∗P < 0.05, ∗∗P < 0.01, and ∗∗∗P < 0.001 compared to the control group. n.s., no significance.