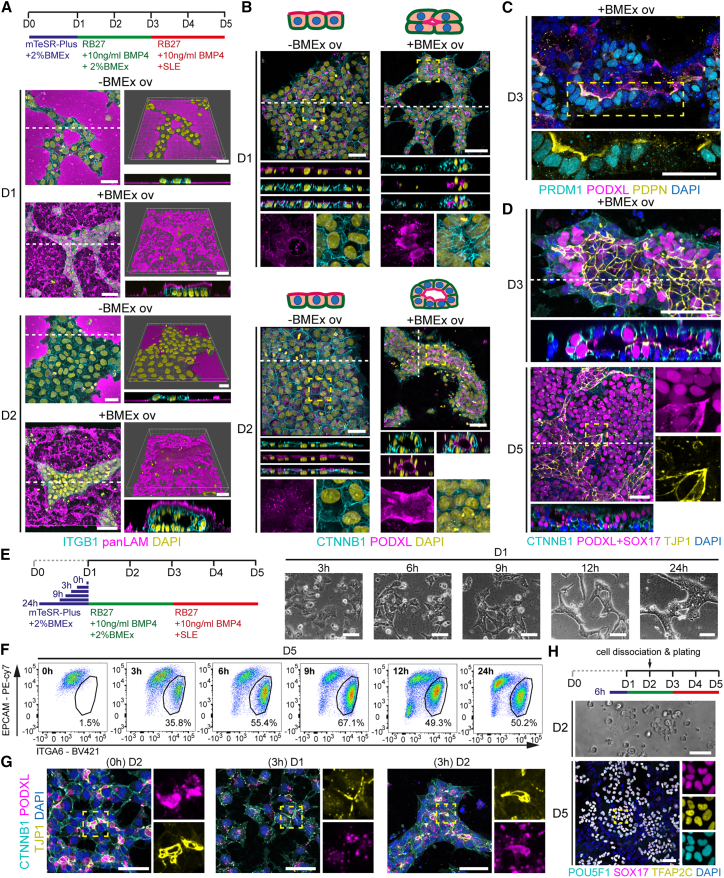
Figure 5.
Differentiation of hPGCLCs is not coupled to lumenogenesis but depends on BMEx overlay
(A) Experimental scheme depicting the conditions used (top) and immunofluorescence for ITGB1 and panLAM as MIP (left) and surface render image (top right) at days 1 and 2, with or without BMEx overlay. Dashed line in left panel shows the level of the digital cross section (bottom right). Scale bars: 50 μm.
(B) Immunofluorescence for CTNNB1 and PODXL at days 1 and 2 with or without BMEx overlay. White dashed line shows the level of the digital cross section (middle panels), and the yellow dashed box is magnified (bottom), showing separated channels. Scale bars: 50 μm.
(C) Immunofluorescence for PRDM1, PODXL, and PDPN at day 3 with BMEx overlay. Dashed box is magnified (bottom), showing separated channels. Scale bar: 50 μm.
(D) Immunofluorescence for CTNNB1, PODXL+SOX17, and TJP1 at days 3 and 5 with BMEx overlay. White dashed line shows the level of the digital cross section (bottom), and the yellow dashed box is magnified (right), showing separated channels. Scale bars: 50 μm.
(E) Experimental scheme depicting the different conditions tested in (F) and (G) (left) and associated bright-field images at day 1 with BMEx overlay. Scale bars: 50 μm.
(F) FACS plots depicting the percentage of double EPCAM+ITGA6+ cells at day 5 in line M54 to test different priming periods.
(G) Immunofluorescence for CTNNB1, PODXL, and TJP1 at days 1 and 2 to test different priming periods. Dashed box is magnified (right), showing separated channels. Scale bars: 30 μm.
(H) Experimental scheme depicting the conditions used to disrupt the lumens at day 2 (top), associated bright-field image at day 2 after dissociation, and immunofluorescence for POU5F1, SOX17, and TFAP2C at day 5. Dashed box is magnified (right), showing separated channels. Scale bars: 50 μm.
See also Figure S3.