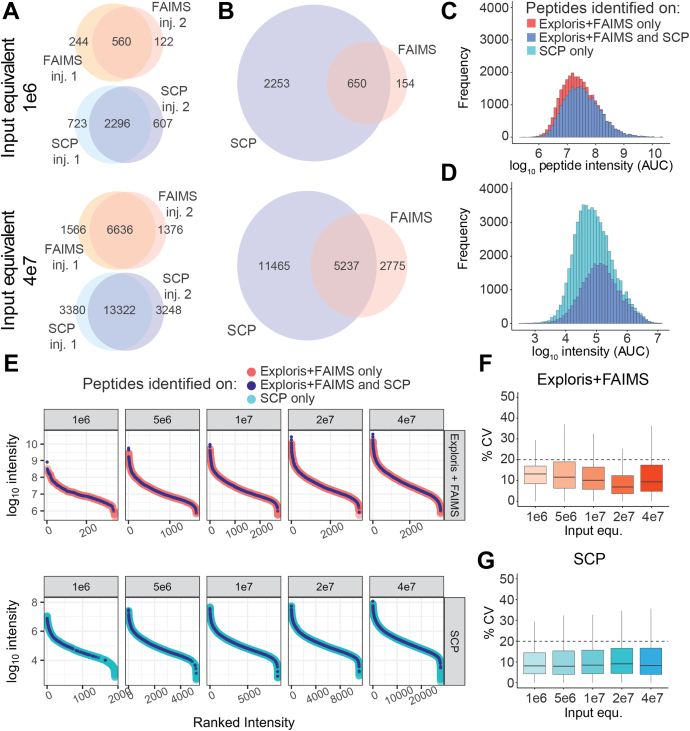
Fig. 3.
HLA-I peptide identifications on the timsTOF single-cell proteomics (SCP) show high reproducibility and extended dynamic range.A, overlap of HLA-I peptides across multiple injections on the Exploris + FAIMS or the timsTOF SCP at 1e6 or 4e7 A375 cell input equivalents from bulk sample preparation. B, overlap of HLA-I peptides between the Exploris + FAIMS and the timsTOF SCP at 1e6 (upper panel) or 4e7 (lower panel) A375 cells. C, Log10 intensity (MS1 AUC) distribution of HLA-I peptides uniquely identified on the Exploris + FAIMS (pink) or both instruments (dark blue). D, Log10 intensity (MS1 AUC) distribution of HLA-I peptides uniquely identified on the timsTOF SCP (light blue) or both instruments (dark blue). E, ranked peptide intensity at different cell input equivalents from bulk sample preparation on the Exploris + FAIMS (upper panel) or the timsTOF SCP (lower panel). Respective intensity of peptides uniquely identified on the Exploris + FAIMS (pink), the timsTOF SCP (light blue), or both instruments (dark blue). F, percent CV of peptide intensity between technical replicates at different cell input equivalents from bulk sample preparation on Exploris + FAIMS. Dashed line indicates 20% CV. G, % CV of peptide intensity between technical replicates identified on the timsTOF SCP at indicated input equivalents from bulk sample preparation. Dashed line indicates 20% CV. AUC, area under the curve; FAIMS, field asymmetric waveform ion mobility spectrometry; HLA, human leukocyte antigen; MS, mass spectrometry.