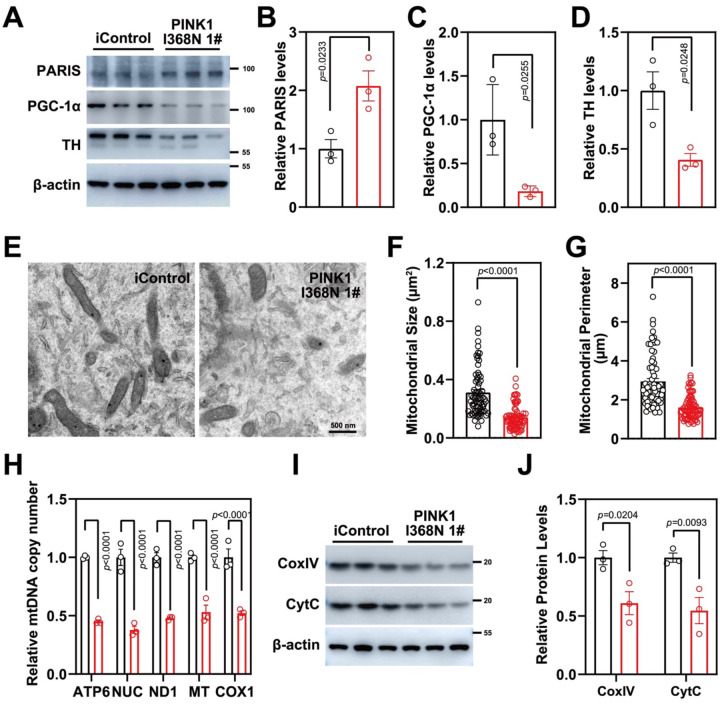
Figure 4. Mitochondrial biogenesis defects in PINK1 p.I368N-1# mutant neurons.
A. Analysis of PARIS, PGC-1α and TH protein level in isogenic control and PINK1 p.I368N 1# mutant neurons by immunoblot.
C. Quantification of immunoblot of PARIS normalized to β-actin. N=3 independent experiments.
D. Quantification of immunoblot of PGC-1α normalized to β-actin. N=3 independent experiments.
E. Quantification of immunoblot of TH normalized to β-actin. N=3 independent experiments.
B. Representative transmission electron microscopy images of neurons from isogenic control and PINK1 p.I368N 1# mutant.
C. Quantification of mitochondrial size per cell body in isogenic control and PINK1 p.I368N 1# mutant neurons. N=15–30 TH positive neurons in each group.
D. Quantification of mitochondrial perimeter per cell body in isogenic control and PINK1 p.I368N 1# mutant neurons. N=15–30 TH positive neurons in each group.
F. Relative mitochondrial DNA copy number quantification of ATP6, NUC, ND1, MT, and COX1 in isogenic control and PINK1 p.I368N 1# mutant neurons. N=3 independent experiments.
E. Analysis of CoxIV and CytC protein level in isogenic control and PINK1 p.I368N 1# mutant neurons by immunoblot.
G. Quantification of immunoblot of CoxIV and CytC in isogenic control and PINK1 p.I368N 1# mutant neurons. N=3 independent experiments.