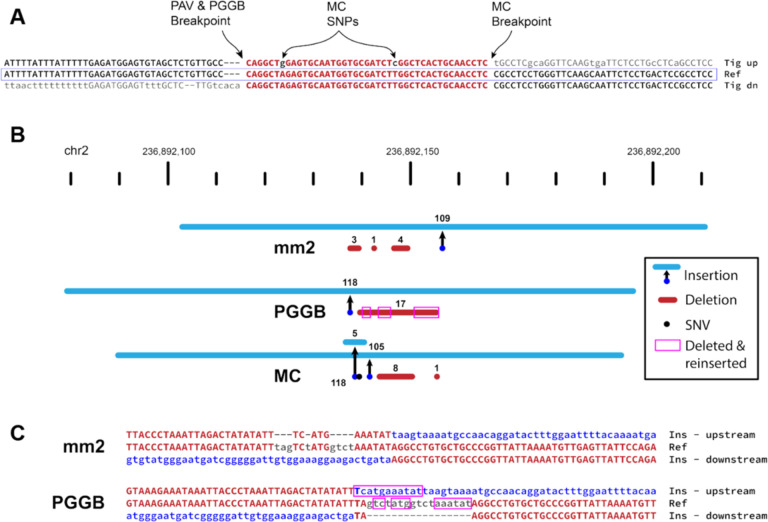
Figure 4: Pangenome graph breakpoints exhibit systematic bias.
(A) A variant with a different breakpoint in the MC graph vs GRCh38 with the red portion showing the imperfect homology between breakpoint placements resulting in two SNPs called in the MC graph in all HPRC haplotypes (A>G and T>C). (B) An SV insertion (blue) paired with deleted bases (red) yields a 101 bp net gain by minimap2 (mm2, HGSVC callset), PGGB, and MC. minimap2 calls three small deletions near the insertion breakpoint. PGGB calls one larger deletion, but re-inserts deleted bases (magenta boxes) into the insertion call resulting in a larger SV insertion than minimap2. MC calls two insertions, two deletions, and a mismatch (black dot). (C) Alignments through breakpoints are shown for minimap2 and PGGB. Bases aligned to the reference are shown in red with matches in upper-case, inserted sequences are blue, and deleted bases are gray. The inserted sequence was not found in GRCh38, and likely represents the deletion of ancestral sequence, where the reference contains the derived (deleted) allele.