Extended Data Fig. 8|. BtSULT suppressed T cell migration in vitro and in vivo.
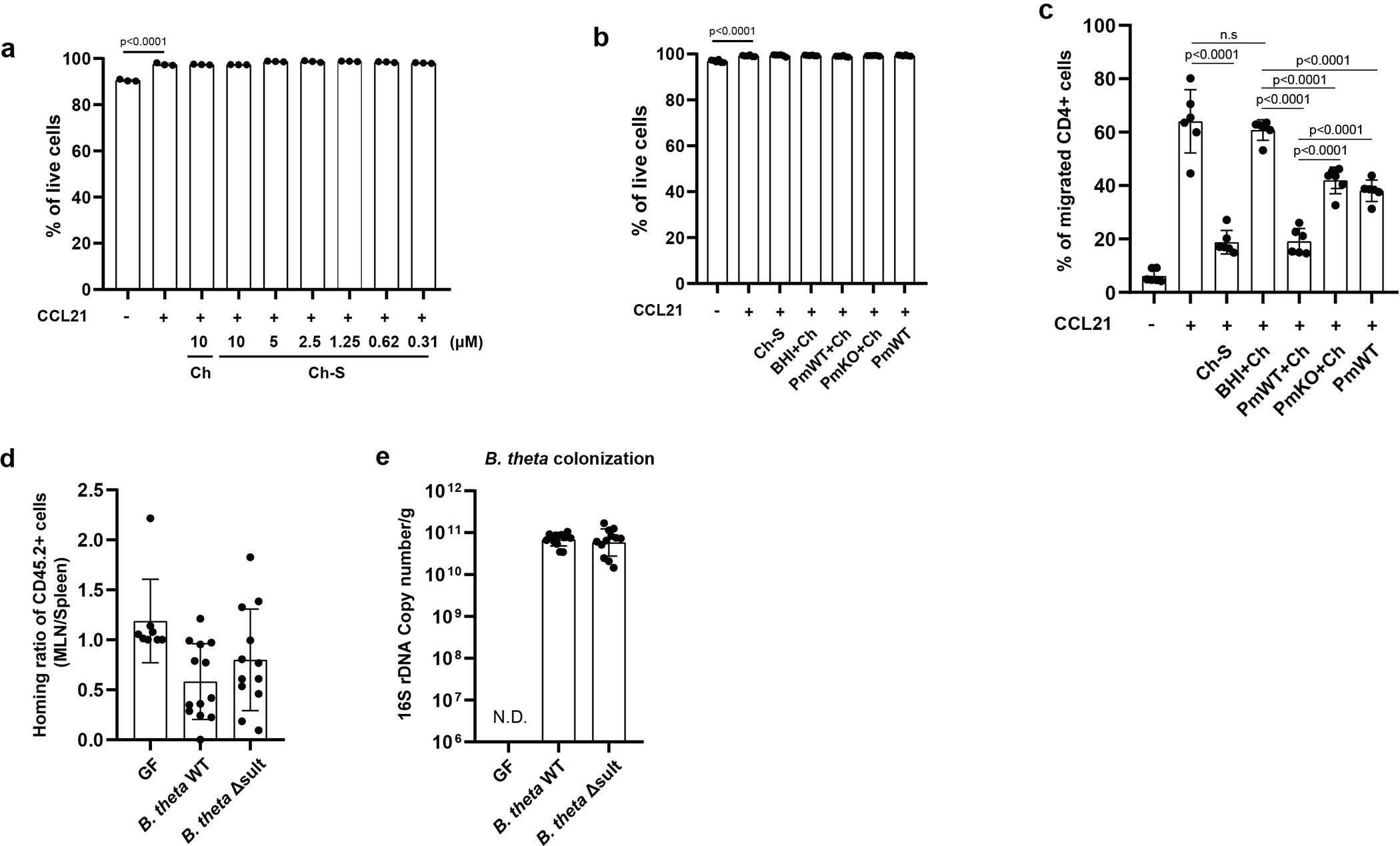
a, Ch-S treatment did not affect leukocyte viability (data correspond to Fig. 4d). Live cell percentage was measured by live/dead dye staining (right) (n=3 biological replicates per condition, data are presented as mean ± S.D, one-way ANOVA followed by Tukey’s multiple comparison test).
b, Bacterial supernatant treatment did not affect leukocyte viability (data correspond to Fig. 4e). Cells were incubated with the following supernatants: BHI+Ch (BHI media + Ch), PmWT+Ch (P. merdae wild-type, cultured with Ch), PmKO+Ch (P. merdae KO cultured with Ch), and PmWT (P. merdae wild-type without Ch) (n=6 biological replicates per condition, data pooled from two independent experiments, data are presented as mean ± S.D., one-way ANOVA followed by Tukey’s multiple comparison test n.s = not significant).
c, Ch-S produced by P. merdae suppressed T cell migration following treatment with CCL21 (200ng/mL). The migrated T cells were identified using CD45, CD90.2 and CD4 antibodies (n=6 biological replicates per condition, data pooled from two independent experiments, data are presented as mean ± S.D., one-way ANOVA followed by Tukey’s multiple comparison test, n.s = not significant).
d, The migration of CCR7-deficient CD45.2+ T cells to MLNs is comparable regardless of recipient mice (data correspond to Fig. 4g). Cells isolated from spleen and MLNs of recipient mice post-24 hours. Results are presented as the normalized ratio of the migrated cells to MLNs divided by the migrated cells to spleens (GF, n=8; B. thetaiotaomicron WT, n=13; B. thetaiotaomicron sult KO, n=12; data pooled from three independent experiments, data are presented as mean ± S.D., one-way ANOVA followed by Tukey’s multiple comparison test).
e, Comparable colonization of B. thetaiotaomicron strains. qPCR analyses for detecting the B. thetaiotaomicron 16S rRNA gene in fecal samples (GF, n=8; B. thetaiotaomicron WT, n=13; B. thetaiotaomicron Δsult, n=12; data pooled from three independent experiments, N.D=not detected).
