Table 1. Data-collection and merging statistics (after anisotropy correction) and structure-refinement parameters for SmTetX.
Values in parentheses are for the highest resolution shell.
| PDB entry | 8aq8 |
| Data collection | |
| X-ray source | BL14.1, BESSY II |
| Wavelength (Å) | 0.9180 |
| Detector | PILATUS 6M |
| Temperature (K) | 100 |
| Crystal-to-detector distance (mm) | 424 |
| No. of oscillation images | 2400 |
| Exposure time per image (s) | 0.1 |
| Oscillation width (°) | 0.1 |
| Data processing | |
| Space group | P21 |
| a, b, c (Å) | 52.9, 160.5, 95.6 |
| α, β, γ (°) | 90, 95.9, 90 |
| Resolution (Å) | 48.21–1.95 (2.01–1.95) |
| No. of reflections | 467117 (25410) |
| No. of unique reflections | 98291 (4915) |
| Multiplicity | 4.8 (5.2) |
| Completeness (spherical) (%) | 85.3 (48.5) |
| Completeness (ellipsoidal) (%) | 96.3 (92.9) |
| R meas | 0.156 (1.702) |
| R p.i.m. | 0.070 (0.729) |
| Mean I/σ(I) | 8.6 (1.1) |
| CC1/2 | 0.997 (0.378) |
| CC* | 0.999 (0.741) |
| Mosaicity (°) | 0.2 |
| Solvent content (%) | 52.6 |
| Refinement | |
| R work | 0.2002 (0.3185) |
| R free (5% reflections) | 0.2410 (0.3289) |
| R all | 0.2022 (0.3175) |
| CCwork | 0.964 (0.592) |
| CCfree | 0.938 (0.617) |
| Mean ADP (Å2) | 28.9 |
| No. of protein chains in asymmetric unit | 4 |
| No. of atoms | 11859 |
| No. of water molecules | 1111 |
| Ligands | 4 × FAD, 6 × ethylene glycol, 2 × polyethylene glycol, 3 ×
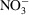 , 2 ×
, 2 ×
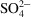 , 4 × Cl−
, 4 × Cl−
|
| R.m.s.d. from ideal bond lengths (Å) | 0.009 |
| R.m.s.d. from ideal angles (°) | 1.498 |
| Ramachandran favoured (%) | 97 |
| Ramachandran outliers (%) | 0 |
| Rama-Z score (Sobolev et al., 2020 ▸) | −1.3 ± 0.2 |
